Identification and functional characterization of soybean root hair microRNAs expressed in response to Bradyrhizobium japonicum infection
- PMID: 25973713
- PMCID: PMC11388829
- DOI: 10.1111/pbi.12387
Identification and functional characterization of soybean root hair microRNAs expressed in response to Bradyrhizobium japonicum infection
Abstract
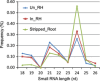
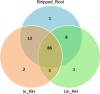
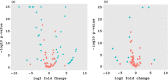
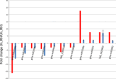
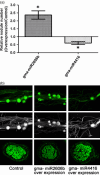
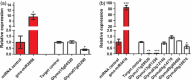
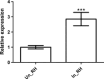
Three soybean [Glycine max (L) Merr.] small RNA libraries were generated and sequenced using the Illumina platform to examine the role of miRNAs during soybean nodulation. The small RNA libraries were derived from root hairs inoculated with Bradyrhizobium japonicum (In_RH) or mock-inoculated with water (Un_RH), as well as from the comparable inoculated stripped root samples (i.e. inoculated roots with the root hairs removed). Sequencing of these libraries identified a total of 114 miRNAs, including 22 novel miRNAs. A comparison of miRNA abundance among the 114 miRNAs identified 66 miRNAs that were differentially expressed between root hairs and stripped roots, and 48 miRNAs that were differentially regulated in infected root hairs in response to B. japonicum when compared to uninfected root hairs (P ≤ 0.05). A parallel analysis of RNA ends (PARE) library was constructed and sequenced to reveal a total of 405 soybean miRNA targets, with most predicted to encode transcription factors or proteins involved in protein modification, protein degradation and hormone pathways. The roles of gma-miR4416 and gma-miR2606b during nodulation were further analysed. Ectopic expression of these two miRNAs in soybean roots resulted in significant changes in nodule numbers. miRNA target information suggested that gma-miR2606b regulates a Mannosyl-oligosaccharide 1, 2-alpha-mannosidase gene, while gma-miR4416 regulates the expression of a rhizobium-induced peroxidase 1 (RIP1)-like peroxidase gene, GmRIP1, during nodulation.
Keywords: gma-miR2606b; gma-miR4416; miRNA; nodulation; root hair; soybean.
© 2015 Society for Experimental Biology, Association of Applied Biologists and John Wiley & Sons Ltd.
Figures
Similar articles
-
Quantitative phosphoproteomic analysis of soybean root hairs inoculated with Bradyrhizobium japonicum.Mol Cell Proteomics. 2012 Nov;11(11):1140-55. doi: 10.1074/mcp.M112.018028. Epub 2012 Jul 25. Mol Cell Proteomics. 2012. PMID: 22843990 Free PMC article.
-
RNA-Seq analysis of nodule development at five different developmental stages of soybean (Glycine max) inoculated with Bradyrhizobium japonicum strain 113-2.Sci Rep. 2017 Feb 7;7:42248. doi: 10.1038/srep42248. Sci Rep. 2017. PMID: 28169364 Free PMC article.
-
Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum.Plant Physiol. 2010 Aug;153(4):1808-22. doi: 10.1104/pp.110.157800. Epub 2010 Jun 9. Plant Physiol. 2010. PMID: 20534735 Free PMC article.
-
Functional genomics dissection of the nodulation autoregulation pathway (AON) in soybean (Glycine max).J Integr Plant Biol. 2025 Mar;67(3):762-772. doi: 10.1111/jipb.13898. Epub 2025 Mar 24. J Integr Plant Biol. 2025. PMID: 40125797 Review.
-
Natural antisense transcripts in plants: a review and identification in soybean infected with Phakopsora pachyrhizi SuperSAGE library.ScientificWorldJournal. 2013 Jun 26;2013:219798. doi: 10.1155/2013/219798. Print 2013. ScientificWorldJournal. 2013. PMID: 23878522 Free PMC article. Review.
Cited by
-
Regulation of Small RNAs and Corresponding Targets in Nod Factor-Induced Phaseolus vulgaris Root Hair Cells.Int J Mol Sci. 2016 Jun 4;17(6):887. doi: 10.3390/ijms17060887. Int J Mol Sci. 2016. PMID: 27271618 Free PMC article.
-
A Multi-Level Iterative Bi-Clustering Method for Discovering miRNA Co-regulation Network of Abiotic Stress Tolerance in Soybeans.Front Plant Sci. 2022 Apr 7;13:860791. doi: 10.3389/fpls.2022.860791. eCollection 2022. Front Plant Sci. 2022. PMID: 35463453 Free PMC article.
-
Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots.Sci Rep. 2017 Feb 6;7:42094. doi: 10.1038/srep42094. Sci Rep. 2017. PMID: 28165059 Free PMC article.
-
Legumes display common and host-specific responses to the rhizobial cellulase CelC2 during primary symbiotic infection.Sci Rep. 2019 Sep 25;9(1):13907. doi: 10.1038/s41598-019-50337-3. Sci Rep. 2019. PMID: 31554862 Free PMC article.
-
Gene Expression in Nitrogen-Fixing Symbiotic Nodule Cells in Medicago truncatula and Other Nodulating Plants.Plant Cell. 2020 Jan;32(1):42-68. doi: 10.1105/tpc.19.00494. Epub 2019 Nov 11. Plant Cell. 2020. PMID: 31712407 Free PMC article. Review.
References
-
- Allen, E. , Xie, Z. , Gustafson, A.M. and Carrington, J.C. (2005) microRNA‐directed phasing during trans‐acting siRNA biogenesis in plants. Cell, 121, 207–221. - PubMed
-
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B, 57, 289–300.
-
- Boualem, A. , Laporte, P. , Jovanovic, M. , Laffont, C. , Plet, J. , Combier, J.P. , Niebel, A. , Crespi, M. and Frugier, F. (2008) MicroRNA166 controls root and nodule development in Medicago truncatula . Plant J. 54, 876–887. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

