Est10: A Novel Alkaline Esterase Isolated from Bovine Rumen Belonging to the New Family XV of Lipolytic Enzymes
- PMID: 25973851
- PMCID: PMC4431682
- DOI: 10.1371/journal.pone.0126651
Est10: A Novel Alkaline Esterase Isolated from Bovine Rumen Belonging to the New Family XV of Lipolytic Enzymes
Abstract
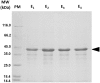
A metagenomic fosmid library from bovine rumen was used to identify clones with lipolytic activity. One positive clone was isolated. The gene responsible for the observed phenotype was identified by in vitro transposon mutagenesis and sequencing and was named est10. The 367 amino acids sequence harbors a signal peptide, the conserved secondary structure arrangement of alpha/beta hydrolases, and a GHSQG pentapeptide which is characteristic of esterases and lipases. Homology based 3D-modelling confirmed the conserved spatial orientation of the serine in a nucleophilic elbow. By sequence comparison, Est10 is related to hydrolases that are grouped into the non-specific Pfam family DUF3089 and to other characterized esterases that were recently classified into the new family XV of lipolytic enzymes. Est10 was heterologously expressed in Escherichia coli as a His-tagged fusion protein, purified and biochemically characterized. Est10 showed maximum activity towards C4 aliphatic chains and undetectable activity towards C10 and longer chains which prompted its classification as an esterase. However, it was able to efficiently catalyze the hydrolysis of aryl esters such as methyl phenylacetate and phenyl acetate. The optimum pH of this enzyme is 9.0, which is uncommon for esterases, and it exhibits an optimal temperature at 40 °C. The activity of Est10 was inhibited by metal ions, detergents, chelating agents and additives. We have characterized an alkaline esterase produced by a still unidentified bacterium belonging to a recently proposed new family of esterases.
Conflict of interest statement
Figures




References
-
- Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzyme and Microbial Technology. 2006;39(2):235–51. 10.1016/j.enzmictec.2005.10.016 - DOI
-
- Jaeger KE, Eggert T. Lipases for biotechnology. Curr Opin Biotechnol. 2002;13(4):390–7. Epub 2002/09/27. . - PubMed
-
- Bornscheuer UT. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS microbiology reviews. 2002;26(1):73–81. Epub 2002/05/15. . - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

