Effort versus Reward: Preparing Samples for Fungal Community Characterization in High-Throughput Sequencing Surveys of Soils
- PMID: 25974078
- PMCID: PMC4431839
- DOI: 10.1371/journal.pone.0127234
Effort versus Reward: Preparing Samples for Fungal Community Characterization in High-Throughput Sequencing Surveys of Soils
Abstract
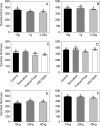
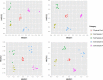
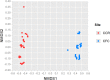
Next generation fungal amplicon sequencing is being used with increasing frequency to study fungal diversity in various ecosystems; however, the influence of sample preparation on the characterization of fungal community is poorly understood. We investigated the effects of four procedural modifications to library preparation for high-throughput sequencing (HTS). The following treatments were considered: 1) the amount of soil used in DNA extraction, 2) the inclusion of additional steps (freeze/thaw cycles, sonication, or hot water bath incubation) in the extraction procedure, 3) the amount of DNA template used in PCR, and 4) the effect of sample pooling, either physically or computationally. Soils from two different ecosystems in Minnesota, USA, one prairie and one forest site, were used to assess the generality of our results. The first three treatments did not significantly influence observed fungal OTU richness or community structure at either site. Physical pooling captured more OTU richness compared to individual samples, but total OTU richness at each site was highest when individual samples were computationally combined. We conclude that standard extraction kit protocols are well optimized for fungal HTS surveys, but because sample pooling can significantly influence OTU richness estimates, it is important to carefully consider the study aims when planning sampling procedures.
Conflict of interest statement
Figures


References
-
- Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88: 1354–64. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

