Antifungal activity of the ribosome-inactivating protein BE27 from sugar beet (Beta vulgaris L.) against the green mould Penicillium digitatum
- PMID: 25976013
- PMCID: PMC6638414
- DOI: 10.1111/mpp.12278
Antifungal activity of the ribosome-inactivating protein BE27 from sugar beet (Beta vulgaris L.) against the green mould Penicillium digitatum
Abstract
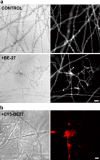
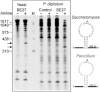
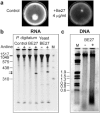
The ribosome-inactivating protein BE27 from sugar beet (Beta vulgaris L.) leaves is an apoplastic protein induced by signalling compounds, such as hydrogen peroxide and salicylic acid, which has been reported to be involved in defence against viruses. Here, we report that, at a concentration much lower than that present in the apoplast, BE27 displays antifungal activity against the green mould Penicillium digitatum, a necrotrophic fungus that colonizes wounds and grows in the inter- and intracellular spaces of the tissues of several edible plants. BE27 is able to enter into the cytosol and kill fungal cells, thus arresting the growth of the fungus. The mechanism of action seems to involve ribosomal RNA (rRNA) N-glycosylase activity on the sarcin-ricin loop of the major rRNA which inactivates irreversibly the fungal ribosomes, thus inhibiting protein synthesis. We compared the C-terminus of the BE27 structure with antifungal plant defensins and hypothesize that a structural motif composed of an α-helix and a β-hairpin, similar to the γ-core motif of defensins, might contribute to the specific interaction with the fungal plasma membranes, allowing the protein to enter into the cell.
Keywords: apoplast; defensin; green mould; plant defence; pokeweed antiviral protein (PAP); polynucleotide:adenosine glycosylase; rRNA N-glycosylase.
© 2015 BSPP AND JOHN WILEY & SONS LTD.
Figures


References
-
- Barbieri, L. , Polito, L. , Bolognesi, A. , Ciani, M. , Pelosi, E. , Farini, V. , Jha, A.K. , Sharma, N. , Vivanco, J.M. , Chambery, A. , Parente, A. and Stirpe, F. (2006) Ribosome‐inactivating proteins in edible plants and purification and characterization of a new ribosome‐inactivating protein from Cucurbita moschata . Biochim. Biophys. Acta, 1760, 783–792. - PubMed
-
- Baykal, U. and Tumer, N.E. (2007) The C‐terminus of pokeweed antiviral protein has distinct roles in transport to the cytosol, ribosome depurination and cytotoxicity. Plant J. 49, 995–1007. - PubMed
-
- Becker, N. and Benhar, I. (2012) Antibody‐based immunotoxins for the treatment of cancer. Antibodies, 1, 39–69.
-
- Berglund, L. , Brunstedt, J. , Nielsen, K.K. , Chen, Z. , Mikkelsen, J.D. and Marcker, K.A. (1995) A proline‐rich chitinase from Beta vulgaris . Plant Mol. Biol. 27, 211–216. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

