BCR repertoire sequencing: different patterns of B-cell activation after two Meningococcal vaccines
- PMID: 25976772
- PMCID: PMC4551417
- DOI: 10.1038/icb.2015.57
BCR repertoire sequencing: different patterns of B-cell activation after two Meningococcal vaccines
Abstract
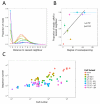
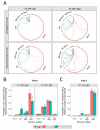
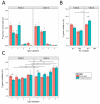
Next-generation sequencing was used to investigate the B-cell receptor heavy chain transcript repertoire of different B-cell subsets (naive, marginal zone (MZ), immunoglobulin M (IgM) memory and IgG memory) at baseline, and of plasma cells (PCs) 7 days following administration of serogroup ACWY meningococcal polysaccharide and protein-polysaccharide conjugate vaccines. Baseline B-cell subsets could be distinguished from each other using a small number of repertoire properties (clonality, mutation from germline and complementarity-determining region 3 (CDR3) length) that were conserved between individuals. However, analyzing the CDR3 amino-acid sequence (which is particularly important for antigen binding) of the baseline subsets showed few sequences shared between individuals. In contrast, day 7 PCs demonstrated nearly 10-fold greater sequence sharing between individuals than the baseline subsets, consistent with the PCs being induced by the vaccine antigen and sharing specificity for a more limited range of epitopes. By annotating PC sequences based on IgG subclass usage and mutation, and also comparing them with the sequences of the baseline cell subsets, we were able to identify different signatures after the polysaccharide and conjugate vaccines. PCs produced after conjugate vaccination were predominantly IgG1, and most related to IgG memory cells. In contrast, after polysaccharide vaccination, the PCs were predominantly IgG2, less mutated and were equally likely to be related to MZ, IgM memory or IgG memory cells. High-throughput B-cell repertoire sequencing thus provides a unique insight into patterns of B-cell activation not possible from more conventional measures of immunogenicity.
Figures



References
-
- Galson JD, Pollard AJ, Trück J, Kelly DF. Studying the antibody repertoire after vaccination: practical applications. Trends Immunol. 2014;35:319–331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

