Longitudinal epigenetic and gene expression profiles analyzed by three-component analysis reveal down-regulation of genes involved in protein translation in human aging
- PMID: 25977295
- PMCID: PMC4551908
- DOI: 10.1093/nar/gkv473
Longitudinal epigenetic and gene expression profiles analyzed by three-component analysis reveal down-regulation of genes involved in protein translation in human aging
Abstract
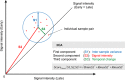
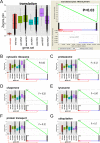
Data on biological mechanisms of aging are mostly obtained from cross-sectional study designs. An inherent disadvantage of this design is that inter-individual differences can mask small but biologically significant age-dependent changes. A serially sampled design (same individual at different time points) would overcome this problem but is often limited by the relatively small numbers of available paired samples and the statistics being used. To overcome these limitations, we have developed a new vector-based approach, termed three-component analysis, which incorporates temporal distance, signal intensity and variance into one single score for gene ranking and is combined with gene set enrichment analysis. We tested our method on a unique age-based sample set of human skin fibroblasts and combined genome-wide transcription, DNA methylation and histone methylation (H3K4me3 and H3K27me3) data. Importantly, our method can now for the first time demonstrate a clear age-dependent decrease in expression of genes coding for proteins involved in translation and ribosome function. Using analogies with data from lower organisms, we propose a model where age-dependent down-regulation of protein translation-related components contributes to extend human lifespan.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Stress-associated H3K4 methylation accumulates during postnatal development and aging of rhesus macaque brain.Aging Cell. 2012 Dec;11(6):1055-64. doi: 10.1111/acel.12007. Epub 2012 Oct 18. Aging Cell. 2012. PMID: 22978322
-
Age-dependent clinical prognostic value of histone modifications in colorectal cancer.Transl Res. 2015 May;165(5):578-88. doi: 10.1016/j.trsl.2014.11.001. Epub 2014 Nov 13. Transl Res. 2015. PMID: 25488396
-
Krebs cycle intermediates regulate DNA and histone methylation: epigenetic impact on the aging process.Ageing Res Rev. 2014 Jul;16:45-65. doi: 10.1016/j.arr.2014.05.004. Epub 2014 Jun 5. Ageing Res Rev. 2014. PMID: 24910305 Review.
-
Reduced DNA methylation patterning and transcriptional connectivity define human skin aging.Aging Cell. 2016 Jun;15(3):563-71. doi: 10.1111/acel.12470. Epub 2016 Mar 23. Aging Cell. 2016. PMID: 27004597 Free PMC article.
-
Reconfiguration of DNA methylation in aging.Mech Ageing Dev. 2015 Nov;151:60-70. doi: 10.1016/j.mad.2015.02.002. Epub 2015 Feb 20. Mech Ageing Dev. 2015. PMID: 25708826 Review.
Cited by
-
Positive and negative feedback regulation of the TGF-β1 explains two equilibrium states in skin aging.iScience. 2024 Apr 10;27(5):109708. doi: 10.1016/j.isci.2024.109708. eCollection 2024 May 17. iScience. 2024. PMID: 38706856 Free PMC article.
-
Longitudinal profiling of the blood transcriptome in an African green monkey aging model.Aging (Albany NY). 2020 Dec 3;13(1):846-864. doi: 10.18632/aging.202190. Epub 2020 Dec 3. Aging (Albany NY). 2020. PMID: 33290253 Free PMC article.
-
Autoimmunomic Signatures of Aging and Age-Related Neurodegenerative Diseases Are Associated With Brain Function and Ribosomal Proteins.Front Aging Neurosci. 2021 May 28;13:679688. doi: 10.3389/fnagi.2021.679688. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34122052 Free PMC article.
-
Predicting age from the transcriptome of human dermal fibroblasts.Genome Biol. 2018 Dec 20;19(1):221. doi: 10.1186/s13059-018-1599-6. Genome Biol. 2018. PMID: 30567591 Free PMC article.
-
Genetic regulation of gene expression and splicing during a 10-year period of human aging.Genome Biol. 2019 Nov 4;20(1):230. doi: 10.1186/s13059-019-1840-y. Genome Biol. 2019. PMID: 31684996 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

