NPDock: a web server for protein-nucleic acid docking
- PMID: 25977296
- PMCID: PMC4489298
- DOI: 10.1093/nar/gkv493
NPDock: a web server for protein-nucleic acid docking
Abstract
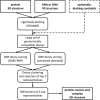
Protein-RNA and protein-DNA interactions play fundamental roles in many biological processes. A detailed understanding of these interactions requires knowledge about protein-nucleic acid complex structures. Because the experimental determination of these complexes is time-consuming and perhaps futile in some instances, we have focused on computational docking methods starting from the separate structures. Docking methods are widely employed to study protein-protein interactions; however, only a few methods have been made available to model protein-nucleic acid complexes. Here, we describe NPDock (Nucleic acid-Protein Docking); a novel web server for predicting complexes of protein-nucleic acid structures which implements a computational workflow that includes docking, scoring of poses, clustering of the best-scored models and refinement of the most promising solutions. The NPDock server provides a user-friendly interface and 3D visualization of the results. The smallest set of input data consists of a protein structure and a DNA or RNA structure in PDB format. Advanced options are available to control specific details of the docking process and obtain intermediate results. The web server is available at http://genesilico.pl/NPDock.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Lukong K.E., Chang K.W., Khandjian E.W., Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. - PubMed
-
- Doudna J.A. Structural genomics of RNA. Nat. Struct. Biol. 2000;7:954–956. - PubMed
-
- Rodrigues J.P., Bonvin A.M. Integrative computational modeling of protein interactions. FEBS J. 2014;281:1988–2003. - PubMed
-
- Wichadakul D., McDermott J., Samudrala R. Prediction and integration of regulatory and protein-protein interactions. Methods Mol. Biol. 2009;541:101–143. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

