Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer
- PMID: 25977370
- PMCID: PMC4456397
- DOI: 10.1126/science.aab0015
Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer
Abstract
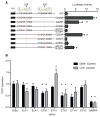
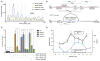
Reactivation of telomerase reverse transcriptase (TERT) expression enables cells to overcome replicative senescence and escape apoptosis, which are fundamental steps in the initiation of human cancer. Multiple cancer types, including up to 83% of glioblastomas (GBMs), harbor highly recurrent TERT promoter mutations of unknown function but specific to two nucleotide positions. We identified the functional consequence of these mutations in GBMs to be recruitment of the multimeric GA-binding protein (GABP) transcription factor specifically to the mutant promoter. Allelic recruitment of GABP is consistently observed across four cancer types, highlighting a shared mechanism underlying TERT reactivation. Tandem flanking native E26 transformation-specific motifs critically cooperate with these mutations to activate TERT, probably by facilitating GABP heterotetramer binding. GABP thus directly links TERT promoter mutations to aberrant expression in multiple cancers.
Copyright © 2015, American Association for the Advancement of Science.
Figures

References
-
- Bryan TM, Cech TR. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. - PubMed
-
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell. 1985;43:405–413. - PubMed
-
- Weinrich SLS, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. - PubMed
-
- Phd PCB, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncology. 2013;14:534–542. - PubMed
-
- Kim NW, et al. Specific Association of Human Telomerase Activity with Immortal Cells and Cancer. Science. 1994;266:2011–2015. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA118816/CA/NCI NIH HHS/United States
- R01CA169316-01/CA/NCI NIH HHS/United States
- R01 CA163336/CA/NCI NIH HHS/United States
- P50 CA097257/CA/NCI NIH HHS/United States
- R25CA112355/CA/NCI NIH HHS/United States
- T32 GM008568/GM/NIGMS NIH HHS/United States
- R01 CA052689/CA/NCI NIH HHS/United States
- P01CA118816-06/CA/NCI NIH HHS/United States
- R01 CA169316/CA/NCI NIH HHS/United States
- R25 CA112355/CA/NCI NIH HHS/United States
- P50CA097257/CA/NCI NIH HHS/United States
- DP2 GM105453/GM/NIGMS NIH HHS/United States
- R01CA163336/CA/NCI NIH HHS/United States
- R01CA52689/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

