Modes of Escherichia coli Dps Interaction with DNA as Revealed by Atomic Force Microscopy
- PMID: 25978038
- PMCID: PMC4433220
- DOI: 10.1371/journal.pone.0126504
Modes of Escherichia coli Dps Interaction with DNA as Revealed by Atomic Force Microscopy
Abstract
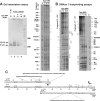
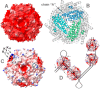
Multifunctional protein Dps plays an important role in iron assimilation and a crucial role in bacterial genome packaging. Its monomers form dodecameric spherical particles accumulating ~400 molecules of oxidized iron ions within the protein cavity and applying a flexible N-terminal ends of each subunit for interaction with DNA. Deposition of iron is a well-studied process by which cells remove toxic Fe2+ ions from the genetic material and store them in an easily accessible form. However, the mode of interaction with linear DNA remained mysterious and binary complexes with Dps have not been characterized so far. It is widely believed that Dps binds DNA without any sequence or structural preferences but several lines of evidence have demonstrated its ability to differentiate gene expression, which assumes certain specificity. Here we show that Dps has a different affinity for the two DNA fragments taken from the dps gene regulatory region. We found by atomic force microscopy that Dps predominantly occupies thermodynamically unstable ends of linear double-stranded DNA fragments and has high affinity to the central part of the branched DNA molecule self-assembled from three single-stranded oligonucleotides. It was proposed that Dps prefers binding to those regions in DNA that provide more contact pads for the triad of its DNA-binding bundle associated with one vertex of the protein globule. To our knowledge, this is the first study revealed the nucleoid protein with an affinity to branched DNA typical for genomic regions with direct and inverted repeats. As a ubiquitous feature of bacterial and eukaryotic genomes, such structural elements should be of particular care, but the protein system evolutionarily adapted for this function is not yet known, and we suggest Dps as a putative component of this system.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

