The Novel Gene CRNDE Encodes a Nuclear Peptide (CRNDEP) Which Is Overexpressed in Highly Proliferating Tissues
- PMID: 25978564
- PMCID: PMC4433331
- DOI: 10.1371/journal.pone.0127475
The Novel Gene CRNDE Encodes a Nuclear Peptide (CRNDEP) Which Is Overexpressed in Highly Proliferating Tissues
Abstract
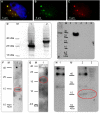
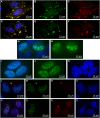
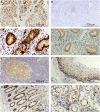
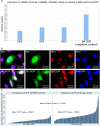
CRNDE, recently described as the lncRNA-coding gene, is overexpressed at RNA level in human malignancies. Its role in gametogenesis, cellular differentiation and pluripotency has been suggested as well. Herein, we aimed to verify our hypothesis that the CRNDE gene may encode a protein product, CRNDEP. By using bioinformatics methods, we identified the 84-amino acid ORF encoded by one of two CRNDE transcripts, previously described by our research team. This ORF was cloned into two expression vectors, subsequently utilized in localization studies in HeLa cells. We also developed a polyclonal antibody against CRNDEP. Its specificity was confirmed in immunohistochemical, cellular localization, Western blot and immunoprecipitation experiments, as well as by showing a statistically significant decrease of endogenous CRNDEP expression in the cells with transient shRNA-mediated knockdown of CRNDE. Endogenous CRNDEP localizes predominantly to the nucleus and its expression seems to be elevated in highly proliferating tissues, like the parabasal layer of the squamous epithelium, intestinal crypts or spermatocytes. After its artificial overexpression in HeLa cells, in a fusion with either the EGFP or DsRed Monomer fluorescent tag, CRNDEP seems to stimulate the formation of stress granules and localize to them. Although the exact role of CRNDEP is unknown, our preliminary results suggest that it may be involved in the regulation of the cell proliferation. Possibly, CRNDEP also participates in oxygen metabolism, considering our in silico results, and the correlation between its enforced overexpression and the formation of stress granules. This is the first report showing the existence of a peptide encoded by the CRNDE gene.
Conflict of interest statement
Figures

References
-
- Szafron L, Lisowska K, Rubel T, Sobiczewski P, Kupryjanczyk J. ING1, PTPN2, PCID2, VGLL1 and LOC388279 genes as potential prognostic markers in ovarian cancer. Int J Mol Med. 2009;24: S40.
-
- Szafron L, Balcerak A, Grzybowska EA, Rubel T, Kupryjanczyk J. The CRNDE, VAV2 and CEBPA genes as new negative prognostic factors in ovarian cancer. Eur. J. Cancer. 2013;49: S215.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

