SIFTER search: a web server for accurate phylogeny-based protein function prediction
- PMID: 25979264
- PMCID: PMC4489292
- DOI: 10.1093/nar/gkv461
SIFTER search: a web server for accurate phylogeny-based protein function prediction
Abstract
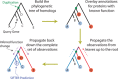
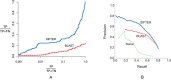
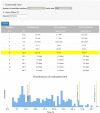
We are awash in proteins discovered through high-throughput sequencing projects. As only a minuscule fraction of these have been experimentally characterized, computational methods are widely used for automated annotation. Here, we introduce a user-friendly web interface for accurate protein function prediction using the SIFTER algorithm. SIFTER is a state-of-the-art sequence-based gene molecular function prediction algorithm that uses a statistical model of function evolution to incorporate annotations throughout the phylogenetic tree. Due to the resources needed by the SIFTER algorithm, running SIFTER locally is not trivial for most users, especially for large-scale problems. The SIFTER web server thus provides access to precomputed predictions on 16 863 537 proteins from 232 403 species. Users can explore SIFTER predictions with queries for proteins, species, functions, and homologs of sequences not in the precomputed prediction set. The SIFTER web server is accessible at http://sifter.berkeley.edu/ and the source code can be downloaded.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

