Porphyromonas gingivalis-derived RgpA-Kgp Complex Activates the Macrophage Urokinase Plasminogen Activator System: IMPLICATIONS FOR PERIODONTITIS
- PMID: 25979345
- PMCID: PMC4481207
- DOI: 10.1074/jbc.M115.645572
Porphyromonas gingivalis-derived RgpA-Kgp Complex Activates the Macrophage Urokinase Plasminogen Activator System: IMPLICATIONS FOR PERIODONTITIS
Abstract
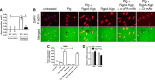
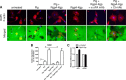
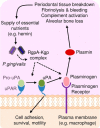
Urokinase plasminogen activator (uPA) converts plasminogen to plasmin, resulting in a proteolytic cascade that has been implicated in tissue destruction during inflammation. Periodontitis is a highly prevalent chronic inflammatory disease characterized by destruction of the tissue and bone that support the teeth. We demonstrate that stimulation of macrophages with the arginine- and lysine-specific cysteine protease complex (RgpA-Kgp complex), produced by the keystone pathogen Porphyromonas gingivalis, dramatically increased their ability to degrade matrix in a uPA-dependent manner. We show that the RgpA-Kgp complex cleaves the inactive zymogens, pro-uPA (at consensus sites Lys(158)-Ile(159) and Lys(135)-Lys(136)) and plasminogen, yielding active uPA and plasmin, respectively. These findings are consistent with activation of the uPA proteolytic cascade by P. gingivalis being required for the pathogen to induce alveolar bone loss in a model of periodontitis and reveal a new host-pathogen interaction in which P. gingivalis activates a critical host proteolytic pathway to promote tissue destruction and pathogen virulence.
Keywords: extracellular matrix; macrophage; periodontal disease; plasmin; plasminogen.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Carmeliet P., Moons L., Lijnen R., Baes M., Lemaître V., Tipping P., Drew A., Eeckhout Y., Shapiro S., Lupu F., Collen D. (1997) Urokinase-generated plasmin activates matrix metalloproteinases during aneurysm formation. Nat. Genet. 17, 439–444 - PubMed
-
- Danø K., Behrendt N., Høyer-Hansen G., Johnsen M., Lund L. R., Ploug M., Rømer J. (2005) Plasminogen activation and cancer. Thromb. Haemost. 93, 676–681 - PubMed
-
- Smith H. W., Marshall C. J. (2010) Regulation of cell signalling by uPAR. Nat. Rev. Mol. Cell Biol. 11, 23–36 - PubMed
-
- Hamilton J. A., Campbell I. K., Wojta J., Cheung D. (1992) Plasminogen activators and their inhibitors in arthritic disease. Ann. N.Y. Acad. Sci. 667, 87–100 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

