Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy
- PMID: 25979999
- PMCID: PMC4507769
- DOI: 10.1093/jxb/erv234
Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy
Abstract
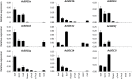
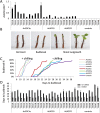
The MADS-domain transcription factor SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) is one of the key integrators of endogenous and environmental signals that promote flowering in the annual species Arabidopsis thaliana. In the deciduous woody perennial vine kiwifruit (Actinidia spp.), environmental signals are integrated to regulate annual cycles of growth and dormancy. Accumulation of chilling during winter is required for dormancy break and flowering in spring. In order to understand the regulation of dormancy and flowering in kiwifruit, nine kiwifruit SOC1-like genes were identified and characterized. All genes affected flowering time of A. thaliana Col-0 and were able to rescue the late flowering phenotype of the soc1-2 mutant when ectopically expressed. A differential capacity for homodimerization was observed, but all proteins were capable of strong interactions with SHORT VEGETATIVE PHASE (SVP) MADS-domain proteins. Largely overlapping spatial domains but distinct expression profiles in buds were identified between the SOC1-like gene family members. Ectopic expression of AcSOC1e, AcSOC1i, and AcSOC1f in Actinidia chinensis had no impact on establishment of winter dormancy and failed to induce precocious flowering, but AcSOC1i reduced the duration of dormancy in the absence of winter chilling. These findings add to our understanding of the SOC1-like gene family and the potential diversification of SOC1 function in woody perennials.
Keywords: Actinidia; budbreak; dormancy; flowering SOC1..
© The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures





Similar articles
-
Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering.J Exp Bot. 2012 Jan;63(2):797-807. doi: 10.1093/jxb/err304. Epub 2011 Nov 9. J Exp Bot. 2012. PMID: 22071267 Free PMC article.
-
Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time.J Exp Bot. 2014 Sep;65(17):4985-95. doi: 10.1093/jxb/eru264. Epub 2014 Jun 19. J Exp Bot. 2014. PMID: 24948678 Free PMC article.
-
Histone modification and activation by SOC1-like and drought stress-related transcription factors may regulate AcSVP2 expression during kiwifruit winter dormancy.Plant Sci. 2019 Apr;281:242-250. doi: 10.1016/j.plantsci.2018.12.001. Epub 2018 Dec 5. Plant Sci. 2019. PMID: 30824057
-
Regulation and function of SOC1, a flowering pathway integrator.J Exp Bot. 2010 May;61(9):2247-54. doi: 10.1093/jxb/erq098. Epub 2010 Apr 22. J Exp Bot. 2010. PMID: 20413527 Review.
-
Make hay when the sun shines: the role of MADS-box genes in temperature-dependant seasonal flowering responses.Plant Sci. 2011 Mar;180(3):447-53. doi: 10.1016/j.plantsci.2010.12.001. Epub 2010 Dec 14. Plant Sci. 2011. PMID: 21421391 Review.
Cited by
-
Overexpression of a SOC1-Related Gene Promotes Bud Break in Ecodormant Poplars.Front Plant Sci. 2021 May 25;12:670497. doi: 10.3389/fpls.2021.670497. eCollection 2021. Front Plant Sci. 2021. PMID: 34113369 Free PMC article.
-
Chromosome-level genome assembly of the diploid blueberry Vaccinium darrowii provides insights into its subtropical adaptation and cuticle synthesis.Plant Commun. 2022 Jul 11;3(4):100307. doi: 10.1016/j.xplc.2022.100307. Epub 2022 Feb 26. Plant Commun. 2022. PMID: 35605198 Free PMC article.
-
StMADS11 Subfamily Gene PfMADS16 From Polypogon fugax Regulates Early Flowering and Seed Development.Front Plant Sci. 2020 May 8;11:525. doi: 10.3389/fpls.2020.00525. eCollection 2020. Front Plant Sci. 2020. PMID: 32457775 Free PMC article.
-
Three FT and multiple CEN and BFT genes regulate maturity, flowering, and vegetative phenology in kiwifruit.J Exp Bot. 2017 Mar 1;68(7):1539-1553. doi: 10.1093/jxb/erx044. J Exp Bot. 2017. PMID: 28369532 Free PMC article.
-
Regulation of wound ethylene biosynthesis by NAC transcription factors in kiwifruit.BMC Plant Biol. 2021 Sep 8;21(1):411. doi: 10.1186/s12870-021-03154-8. BMC Plant Biol. 2021. PMID: 34496770 Free PMC article.
References
-
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara-Silva F, Yanofsky MF. 2000. MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. The Plant Journal 24, 457–466. - PubMed
-
- Bielenberg DG, Wang Y, Li ZG, Zhebentyayeva T, Fan SH, Reighard GL, Scorza R, Abbott AG. 2008. Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genetics and Genomes 4, 495–507.
-
- Borner R, Kampmann G, Chandler J, Gleißner R, Wisman E, Apel K, Melzer S. 2000. A MADS domain gene involved in the transition to flowering in Arabidopsis . The Plant Journal 24, 591–599. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

