A Hoxa13:Cre mouse strain for conditional gene manipulation in developing limb, hindgut, and urogenital system
- PMID: 25980463
- PMCID: PMC4486385
- DOI: 10.1002/dvg.22859
A Hoxa13:Cre mouse strain for conditional gene manipulation in developing limb, hindgut, and urogenital system
Abstract
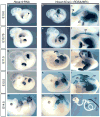
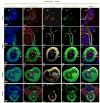
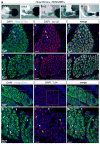
The developing limb is a useful model for studying organogenesis and developmental processes. Although Cre alleles exist for conditional loss- or gain-of-function in limbs, Cre alleles targeting specific limb subdomains are desirable. Here we report on the generation of the Hoxa13:Cre line, in which the Cre gene is inserted in the endogenous Hoxa13 gene. We provide evidence that the Cre is active in embryonic tissues/regions where the endogenous Hoxa13 gene is expressed. Our results show that cells expressing Hoxa13 in developing limb buds contribute to the entire autopod (hand/feet) skeleton and validate Hoxa13 as a distal limb marker as far as the skeleton is concerned. In contrast, in the limb musculature, Cre-based fate mapping shows that almost all muscle masses of the zeugopod (forearm) and part of the triceps contain Hoxa13-expressing cells and/or their descendants. Besides the limb, the activity of the Cre is detectable in the urogenital system and the hindgut, primarily in the epithelium and smooth muscles. Together our data show that the Hoxa13:Cre allele is a useful tool for conditional gene manipulation in the urogenital system, posterior digestive tract, autopod and part of the limb musculature.
Keywords: fate specification; genetics; gut; limb/wing/appendage; mammal; muscle; organism; organogenesis; process; reproductive; skeletal; tissue.
© 2015 Wiley Periodicals, Inc.
Figures




References
-
- Copeland NG, Jenkins NA, Court DL. Recombineering: A powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. - PubMed
-
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. - PubMed
-
- Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, Chambon P. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb auto-pod. Development. 1996;122:2997–3011. - PubMed
-
- Haack H, Gruss P. The establishment of murine Hox-1 expression domains during patterning of the limb. Dev Biol. 1993;157:410–422. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

