A brief review of nucleosome structure
- PMID: 25980611
- PMCID: PMC4598263
- DOI: 10.1016/j.febslet.2015.05.016
A brief review of nucleosome structure
Abstract
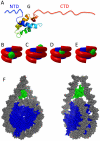
The nucleosomal subunit organization of chromatin provides a multitude of functions. Nucleosomes elicit an initial ∼7-fold linear compaction of genomic DNA. They provide a critical mechanism for stable repression of genes and other DNA-dependent activities by restricting binding of trans-acting factors to cognate DNA sequences. Conversely they are engineered to be nearly meta-stable and disassembled (and reassembled) in a facile manner to allow rapid access to the underlying DNA during processes such as transcription, replication and DNA repair. Nucleosomes protect the genome from DNA damaging agents and provide a lattice onto which a myriad of epigenetic signals are deposited. Moreover, vast strings of nucleosomes provide a framework for assembly of the chromatin fiber and higher-order chromatin structures. Thus, in order to provide a foundation for understanding these functions, we present a review of the basic elements of nucleosome structure and stability, including the association of linker histones.
Keywords: Chromatin structure; Histone; Nucleosome structure.
Copyright © 2015 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. - PubMed
-
- Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–7. - PubMed
-
- van Holde KE. Chromatin. Springer Verlag; New York: 1989.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

