Lineage-Specific Viral Hijacking of Non-canonical E3 Ubiquitin Ligase Cofactors in the Evolution of Vif Anti-APOBEC3 Activity
- PMID: 25981045
- PMCID: PMC4613747
- DOI: 10.1016/j.celrep.2015.04.038
Lineage-Specific Viral Hijacking of Non-canonical E3 Ubiquitin Ligase Cofactors in the Evolution of Vif Anti-APOBEC3 Activity
Abstract
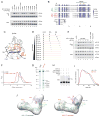
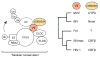
HIV-1 encodes the accessory protein Vif, which hijacks a host Cullin-RING ubiquitin ligase (CRL) complex as well as the non-canonical cofactor CBFβ, to antagonize APOBEC3 antiviral proteins. Non-canonical cofactor recruitment to CRL complexes by viral factors, to date, has only been attributed to HIV-1 Vif. To further study this phenomenon, we employed a comparative approach combining proteomic, biochemical, structural, and virological techniques to investigate Vif complexes across the lentivirus genus, including primate (HIV-1 and simian immunodeficiency virus macaque [SIVmac]) and non-primate (FIV, BIV, and MVV) viruses. We find that CBFβ is completely dispensable for the activity of non-primate lentiviral Vif proteins. Furthermore, we find that BIV Vif requires no cofactor and that MVV Vif requires a novel cofactor, cyclophilin A (CYPA), for stable CRL complex formation and anti-APOBEC3 activity. We propose modular conservation of Vif complexes allows for potential exaptation of functions through the acquisition of non-CRL-associated host cofactors while preserving anti-APOBEC3 activity.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Bosco Da, Eisenmesser EZ, Clarkson MW, Wolf-Watz M, Labeikovsky W, Millet O, Kern D. Dissecting the microscopic steps of the cyclophilin A enzymatic cycle on the biological HIV-1 capsid substrate by NMR. J Mol Biol. 2010;403:723–738. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

