Identification and characterization of parasitism genes from the pinewood nematode Bursaphelenchus xylophilus reveals a multilayered detoxification strategy
- PMID: 25981957
- PMCID: PMC6638532
- DOI: 10.1111/mpp.12280
Identification and characterization of parasitism genes from the pinewood nematode Bursaphelenchus xylophilus reveals a multilayered detoxification strategy
Abstract
The migratory endoparasitic nematode Bursaphelenchus xylophilus, which is the causal agent of pine wilt disease, has phytophagous and mycetophagous phases during its life cycle. This highly unusual feature distinguishes it from other plant-parasitic nematodes and requires profound changes in biology between modes. During the phytophagous stage, the nematode migrates within pine trees, feeding on the contents of parenchymal cells. Like other plant pathogens, B. xylophilus secretes effectors from pharyngeal gland cells into the host during infection. We provide the first description of changes in the morphology of these gland cells between juvenile and adult life stages. Using a comparative transcriptomics approach and an effector identification pipeline, we identify numerous novel parasitism genes which may be important for the mediation of interactions of B. xylophilus with its host. In-depth characterization of all parasitism genes using in situ hybridization reveals two major categories of detoxification proteins, those specifically expressed in either the pharyngeal gland cells or the digestive system. These data suggest that B. xylophilus incorporates effectors in a multilayer detoxification strategy in order to protect itself from host defence responses during phytophagy.
Keywords: Bursaphelenchus xylophilus; effectors; gland cells; transcriptome; xenobiotic metabolism.
© 2015 BSPP AND JOHN WILEY & SONS LTD.
Figures

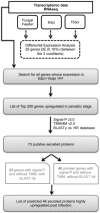


References
-
- Bendtsen, J.D. , Nielsen, H. , von Heijne, G. and Brunak, S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. - PubMed
-
- Carletti, B. , Paoli, F. , Isidoro, N. and Roversi, P.F. (2013) Ultrastructure of the anterior alimentary tract of Bursaphelenchus mucronatus Mamiya et Enda (Nematoda Aphelenchoididae). Redia, XCVI, 69–77.
-
- Chen, Q. , Rehman, S. , Smant, G. and Jones, J.T. (2005) Functional analysis of pathogenicity proteins of the potato cyst nematode Globodera rostochiensis using RNAi. Mol. Plant–Microbe Interact. 18, 621–625. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

