Metagenomics: tools and insights for analyzing next-generation sequencing data derived from biodiversity studies
- PMID: 25983555
- PMCID: PMC4426941
- DOI: 10.4137/BBI.S12462
Metagenomics: tools and insights for analyzing next-generation sequencing data derived from biodiversity studies
Abstract
Advances in next-generation sequencing (NGS) have allowed significant breakthroughs in microbial ecology studies. This has led to the rapid expansion of research in the field and the establishment of "metagenomics", often defined as the analysis of DNA from microbial communities in environmental samples without prior need for culturing. Many metagenomics statistical/computational tools and databases have been developed in order to allow the exploitation of the huge influx of data. In this review article, we provide an overview of the sequencing technologies and how they are uniquely suited to various types of metagenomic studies. We focus on the currently available bioinformatics techniques, tools, and methodologies for performing each individual step of a typical metagenomic dataset analysis. We also provide future trends in the field with respect to tools and technologies currently under development. Moreover, we discuss data management, distribution, and integration tools that are capable of performing comparative metagenomic analyses of multiple datasets using well-established databases, as well as commonly used annotation standards.
Keywords: computational tools; data analysis; metagenomics; next-generation sequencing.
Figures
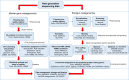
References
-
- Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–52. - PubMed
-
- Tringe SG, von Mering C, Kobayashi A, et al. Comparative metagenomics of microbial communities. Science. 2005;308(5721):554–7. - PubMed
-
- Tringe SSG, Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol. 2008;11(5):442–6. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

