The genetic architecture of growth and fillet traits in farmed Atlantic salmon (Salmo salar)
- PMID: 25985885
- PMCID: PMC4436873
- DOI: 10.1186/s12863-015-0215-y
The genetic architecture of growth and fillet traits in farmed Atlantic salmon (Salmo salar)
Abstract
Background: Performance and quality traits such as harvest weight, fillet weight and flesh color are of economic importance to the Atlantic salmon aquaculture industry. The genetic factors underlying these traits are of scientific and commercial interest. However, such traits are typically polygenic in nature, with the number and size of QTL likely to vary between studies and populations. The aim of this study was to investigate the genetic basis of several growth and fillet traits measured at harvest in a large farmed salmon population by using SNP markers. Due to the marked heterochiasmy in salmonids, an efficient two-stage mapping approach was applied whereby QTL were detected using a sire-based linkage analysis, a sparse SNP marker map and exploiting low rates of recombination, while a subsequent dam-based analysis focused on the significant chromosomes with a denser map to confirm QTL and estimate their position.
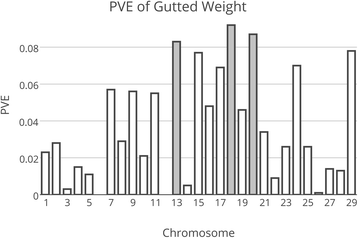
Results: The harvest traits all showed significant heritability, ranging from 0.05 for fillet yield up to 0.53 for the weight traits. In the sire-based analysis, 1695 offspring with trait records and their 20 sires were successfully genotyped for the SNPs on the sparse map. Chromosomes 13, 18, 19 and 20 were shown to harbor genome-wide significant QTL affecting several growth-related traits. The QTL on chr. 13, 18 and 20 were detected in the dam-based analysis using 512 offspring from 10 dams and explained approximately 6-7 % of the within-family variation in these traits.
Conclusions: We have detected several QTL affecting economically important complex traits in a commercial salmon population. Overall, the results suggest that the traits are relatively polygenic and that QTL tend to be pleiotropic (affecting the weight of several components of the harvested fish). Comparison of QTL regions across studies suggests that harvest trait QTL tend to be relatively population-specific. Therefore, the application of marker or genomic selection for improvement in these traits is likely to be most effective when the discovery population is closely related to the selection candidates (e.g. within-family genomic selection).
Figures
References
-
- Gjedrem T, Robinson N, Rye M. The importance of selective breeding in aquaculture to meet future demands for animal protein: A review. Aquaculture. 2012;350–353:117–129. doi: 10.1016/j.aquaculture.2012.04.008. - DOI
-
- Macqueen DJ, Johnston IA. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from spe. Proc R Soc B. 2014;281:20132881. doi: 10.1098/rspb.2013.2881. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

