The biological functions of Naa10 - From amino-terminal acetylation to human disease
- PMID: 25987439
- PMCID: PMC4461483
- DOI: 10.1016/j.gene.2015.04.085
The biological functions of Naa10 - From amino-terminal acetylation to human disease
Abstract
N-terminal acetylation (NTA) is one of the most abundant protein modifications known, and the N-terminal acetyltransferase (NAT) machinery is conserved throughout all Eukarya. Over the past 50 years, the function of NTA has begun to be slowly elucidated, and this includes the modulation of protein-protein interaction, protein-stability, protein function, and protein targeting to specific cellular compartments. Many of these functions have been studied in the context of Naa10/NatA; however, we are only starting to really understand the full complexity of this picture. Roughly, about 40% of all human proteins are substrates of Naa10 and the impact of this modification has only been studied for a few of them. Besides acting as a NAT in the NatA complex, recently other functions have been linked to Naa10, including post-translational NTA, lysine acetylation, and NAT/KAT-independent functions. Also, recent publications have linked mutations in Naa10 to various diseases, emphasizing the importance of Naa10 research in humans. The recent design and synthesis of the first bisubstrate inhibitors that potently and selectively inhibit the NatA/Naa10 complex, monomeric Naa10, and hNaa50 further increases the toolset to analyze Naa10 function.
Keywords: Acetyltransferases; Amino-terminal acetylation; Enzymology; NAA10; NAA15; NAA50; Ogden syndrome; Proteins; Proteomics; ard1.
Copyright © 2015 Elsevier B.V. All rights reserved.
Conflict of interest statement
G.J.L. serves on the medical advisory board of GenePeeks, Inc. and the scientific advisory board of Omicia, Inc. The study did not involve these companies and did not use products from these companies.
Figures




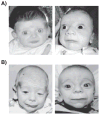
References
-
- Aivaliotis M, Gevaert K, Falb M, Tebbe A, Konstantinidis K, Bisle B, Klein C, Martens L, Staes A, Timmerman E, Van Damme J, Siedler F, Pfeiffer F, Vandekerckhove J, Oesterhelt D. Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis. J Proteome Res. 2007;6:2195–2204. - PubMed
-
- Aksnes H, Hole K, Arnesen T. Molecular, cellular, and physiological significance of N-terminal acetylation. Int Rev Cell Mol Biol. 2015a;316:267–305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

