Genome-wide analysis links NFATC2 with asparaginase hypersensitivity
- PMID: 25987655
- PMCID: PMC4492197
- DOI: 10.1182/blood-2015-02-628800
Genome-wide analysis links NFATC2 with asparaginase hypersensitivity
Abstract
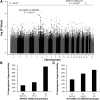
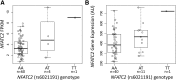
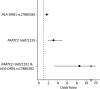
Asparaginase is used to treat acute lymphoblastic leukemia (ALL); however, hypersensitivity reactions can lead to suboptimal asparaginase exposure. Our objective was to use a genome-wide approach to identify loci associated with asparaginase hypersensitivity in children with ALL enrolled on St. Jude Children's Research Hospital (SJCRH) protocols Total XIIIA (n = 154), Total XV (n = 498), and Total XVI (n = 271), or Children's Oncology Group protocols POG 9906 (n = 222) and AALL0232 (n = 2163). Germline DNA was genotyped using the Affymetrix 500K, Affymetrix 6.0, or the Illumina Exome BeadChip array. In multivariate logistic regression, the intronic rs6021191 variant in nuclear factor of activated T cells 2 (NFATC2) had the strongest association with hypersensitivity (P = 4.1 × 10(-8); odds ratio [OR] = 3.11). RNA-seq data available from 65 SJCRH ALL tumor samples and 52 Yoruba HapMap samples showed that samples carrying the rs6021191 variant had higher NFATC2 expression compared with noncarriers (P = 1.1 × 10(-3) and 0.03, respectively). The top ranked nonsynonymous polymorphism was rs17885382 in HLA-DRB1 (P = 3.2 × 10(-6); OR = 1.63), which is in near complete linkage disequilibrium with the HLA-DRB1*07:01 allele we previously observed in a candidate gene study. The strongest risk factors for asparaginase allergy are variants within genes regulating the immune response.
© 2015 by The American Society of Hematology.
Figures

References
-
- Killander D, Dohlwitz A, Engstedt L, et al. Hypersensitive reactions and antibody formation during L-asparaginase treatment of children and adults with acute leukemia. Cancer. 1976;37(1):220–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA 142665/CA/NCI NIH HHS/United States
- U10 CA98413/CA/NCI NIH HHS/United States
- CA 36401/CA/NCI NIH HHS/United States
- R01 CA142665/CA/NCI NIH HHS/United States
- RCA 156449/PHS HHS/United States
- R37 CA036401/CA/NCI NIH HHS/United States
- U10 CA098413/CA/NCI NIH HHS/United States
- U24 CA114766/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- U10 CA098543/CA/NCI NIH HHS/United States
- R01 CA036401/CA/NCI NIH HHS/United States
- GM 92666/GM/NIGMS NIH HHS/United States
- CA 21765/CA/NCI NIH HHS/United States
- U10 CA98543/CA/NCI NIH HHS/United States
- U01 GM092666/GM/NIGMS NIH HHS/United States
- P50 GM115279/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

