Functional loss of IκBε leads to NF-κB deregulation in aggressive chronic lymphocytic leukemia
- PMID: 25987724
- PMCID: PMC4451125
- DOI: 10.1084/jem.20142009
Functional loss of IκBε leads to NF-κB deregulation in aggressive chronic lymphocytic leukemia
Abstract
NF-κB is constitutively activated in chronic lymphocytic leukemia (CLL); however, the implicated molecular mechanisms remain largely unknown. Thus, we performed targeted deep sequencing of 18 core complex genes within the NF-κB pathway in a discovery and validation CLL cohort totaling 315 cases. The most frequently mutated gene was NFKBIE (21/315 cases; 7%), which encodes IκBε, a negative regulator of NF-κB in normal B cells. Strikingly, 13 of these cases carried an identical 4-bp frameshift deletion, resulting in a truncated protein. Screening of an additional 377 CLL cases revealed that NFKBIE aberrations predominated in poor-prognostic patients and were associated with inferior outcome. Minor subclones and/or clonal evolution were also observed, thus potentially linking this recurrent event to disease progression. Compared with wild-type patients, NFKBIE-deleted cases showed reduced IκBε protein levels and decreased p65 inhibition, along with increased phosphorylation and nuclear translocation of p65. Considering the central role of B cell receptor (BcR) signaling in CLL pathobiology, it is notable that IκBε loss was enriched in aggressive cases with distinctive stereotyped BcR, likely contributing to their poor prognosis, and leading to an altered response to BcR inhibitors. Because NFKBIE deletions were observed in several other B cell lymphomas, our findings suggest a novel common mechanism of NF-κB deregulation during lymphomagenesis.
© 2015 Mansouri et al.
Figures
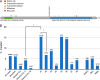

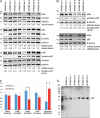

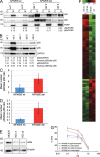
Comment in
-
Augmenting NF-κB in poor-risk CLL: A general paradigm for other cancers?J Exp Med. 2015 Jun 1;212(6):830-1. doi: 10.1084/jem.2126insight4. J Exp Med. 2015. PMID: 26034117 Free PMC article. No abstract available.
References
-
- Agathangelidis A., Darzentas N., Hadzidimitriou A., Brochet X., Murray F., Yan X.J., Davis Z., van Gastel-Mol E.J., Tresoldi C., Chu C.C., et al. 2012. Stereotyped B-cell receptors in one-third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies. Blood. 119:4467–4475. 10.1182/blood-2011-11-393694 - DOI - PMC - PubMed
-
- Alves B.N., Tsui R., Almaden J., Shokhirev M.N., Davis-Turak J., Fujimoto J., Birnbaum H., Ponomarenko J., and Hoffmann A.. 2014. IκBε is a key regulator of B cell expansion by providing negative feedback on cRel and RelA in a stimulus-specific manner. J. Immunol. 192:3121–3132. 10.4049/jimmunol.1302351 - DOI - PMC - PubMed
-
- Baliakas P., Hadzidimitriou A., Sutton L.-A., Minga E., Agathangelidis A., Nichelatti M., Tsanousa A., Scarfò L., Davis Z., Yan X.-J., et al. 2014. Clinical effect of stereotyped B-cell receptor immunoglobulins in chronic lymphocytic leukaemia: a retrospective multicentre study. The Lancet Haematology. 1:e74–e84. 10.1016/S2352-3026(14)00005-2 - DOI - PubMed
-
- Baliakas P., Hadzidimitriou A., Sutton L.A., Rossi D., Minga E., Villamor N., Larrayoz M., Kminkova J., Agathangelidis A., Davis Z., et al. European Research Initiative on CLL (ERIC). 2015. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 29:329–336. 10.1038/leu.2014.196 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

