Pyroglutamyl-N-terminal prion protein fragments in sheep brain following the development of transmissible spongiform encephalopathies
- PMID: 25988175
- PMCID: PMC4429639
- DOI: 10.3389/fmolb.2015.00007
Pyroglutamyl-N-terminal prion protein fragments in sheep brain following the development of transmissible spongiform encephalopathies
Abstract
Protein misfolding, protein aggregation and disruption to cellular proteostasis are key processes in the propagation of disease and, in some progressive neurodegenerative diseases of the central nervous system, the misfolded protein can act as a self-replicating template or prion converting its normal isoform into a misfolded copy of itself. We have investigated the sheep transmissible spongiform encephalopathy, scrapie, and developed a multiple selected reaction monitoring (mSRM) mass spectrometry assay to quantify brain peptides representing the "ragged" N-terminus and the core of ovine prion protein (PrP(Sc)) by using Q-Tof mass spectrometry. This allowed us to identify pyroglutamylated N-terminal fragments of PrP(Sc) at residues 86, 95 and 101, and establish that these fragments were likely to be the result of in vivo processes. We found that the ratios of pyroglutamylated PrP(Sc) fragments were different in sheep of different breeds and geographical origin, and our expanded ovine PrP(Sc) assay was able to determine the ratio and allotypes of PrP accumulating in diseased brain of PrP heterozygous sheep; it also revealed significant differences between N-terminal amino acid profiles (N-TAAPs) in other types of ovine prion disease, CH1641 scrapie and ovine BSE. Variable rates of PrP misfolding, aggregation and degradation are the likely basis for phenotypic (or strain) differences in prion-affected animals and our mass spectrometry-based approach allows the simultaneous investigation of factors such as post-translational modification (pyroglutamyl formation), conformation (by N-TAAP analysis) and amino-acid polymorphisms (allotype ratio) which affect the kinetics of these proteostatic processes.
Keywords: amyloid disease; mass spectrometry; prions; pyroglutamate; strain differentiation.
Figures



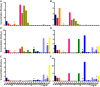
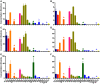

References
-
- Baldwin M. A., Falick A. M., Gibson B. W., Prusiner S. B., Stahl N., Burlingame A. L. (1990). Tandem mass spectrometry of peptides with N-terminal glutamine. Studies on a prion protein peptide. J. Am. Soc. Mass Spectrom. 1, 258–264 10.1016/1044-0305(90)85043-L - DOI
-
- Bruce M. E. (1993). Scrapie strain variation and mutation. Br. Med. Bull. 49, 822–838. - PubMed
-
- Carr S. A., Biemann K. (1984). Identification of posttranslationally modified amino acids in proteins by mass spectrometry, in Methods in Enzymology, eds Wold F., Moldave K. (Orlando, FL: Elsevier; ), 29–58. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

