Heparin affinity purification of extracellular vesicles
- PMID: 25988257
- PMCID: PMC4437317
- DOI: 10.1038/srep10266
Heparin affinity purification of extracellular vesicles
Abstract
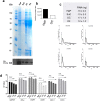
Extracellular vesicles (EVs) are lipid membrane vesicles released by cells. They carry active biomolecules including DNA, RNA, and protein which can be transferred to recipient cells. Isolation and purification of EVs from culture cell media and biofluids is still a major challenge. The most widely used isolation method is ultracentrifugation (UC) which requires expensive equipment and only partially purifies EVs. Previously we have shown that heparin blocks EV uptake in cells, supporting a direct EV-heparin interaction. Here we show that EVs can be purified from cell culture media and human plasma using ultrafiltration (UF) followed by heparin-affinity beads. UF/heparin-purified EVs from cell culture displayed the EV marker Alix, contained a diverse RNA profile, had lower levels of protein contamination, and were functional at binding to and uptake into cells. RNA yield was similar for EVs isolated by UC. We were able to detect mRNAs in plasma samples with comparable levels to UC samples. In conclusion, we have discovered a simple, scalable, and effective method to purify EVs taking advantage of their heparin affinity.
Figures




References
-
- Lee Y., El Andaloussi S. & Wood M. J. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum. Mol. Genet. 21 (R1): R125–34 (2012). - PubMed
-
- Thery C., Ostrowski M. & Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 (2009). - PubMed
-
- Camussi G., Deregibus M. C., Bruno S., Cantaluppi V. & Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 78, 838–848 (2010). - PubMed
-
- Cocucci E., Racchetti G. & Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 19, 43–51 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

