MICCA: a complete and accurate software for taxonomic profiling of metagenomic data
- PMID: 25988396
- PMCID: PMC4649890
- DOI: 10.1038/srep09743
MICCA: a complete and accurate software for taxonomic profiling of metagenomic data
Abstract
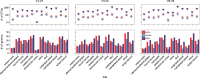
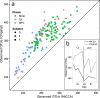
The introduction of high throughput sequencing technologies has triggered an increase of the number of studies in which the microbiota of environmental and human samples is characterized through the sequencing of selected marker genes. While experimental protocols have undergone a process of standardization that makes them accessible to a large community of scientist, standard and robust data analysis pipelines are still lacking. Here we introduce MICCA, a software pipeline for the processing of amplicon metagenomic datasets that efficiently combines quality filtering, clustering of Operational Taxonomic Units (OTUs), taxonomy assignment and phylogenetic tree inference. MICCA provides accurate results reaching a good compromise among modularity and usability. Moreover, we introduce a de-novo clustering algorithm specifically designed for the inference of Operational Taxonomic Units (OTUs). Tests on real and synthetic datasets shows that thanks to the optimized reads filtering process and to the new clustering algorithm, MICCA provides estimates of the number of OTUs and of other common ecological indices that are more accurate and robust than currently available pipelines. Analysis of public metagenomic datasets shows that the higher consistency of results improves our understanding of the structure of environmental and human associated microbial communities. MICCA is an open source project.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

