The Rice High-Affinity Potassium Transporter1;1 Is Involved in Salt Tolerance and Regulated by an MYB-Type Transcription Factor
- PMID: 25991736
- PMCID: PMC4741328
- DOI: 10.1104/pp.15.00298
The Rice High-Affinity Potassium Transporter1;1 Is Involved in Salt Tolerance and Regulated by an MYB-Type Transcription Factor
Abstract
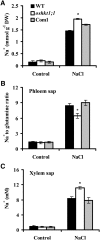
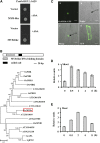
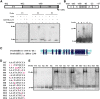
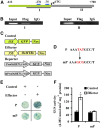
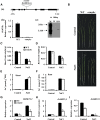
Sodium transporters play key roles in plant tolerance to salt stress. Here, we report that a member of the High-Affinity K(+) Transporter (HKT) family, OsHKT1;1, in rice (Oryza sativa 'Nipponbare') plays an important role in reducing Na(+) accumulation in shoots to cope with salt stress. The oshkt1;1 mutant plants displayed hypersensitivity to salt stress. They contained less Na(+) in the phloem sap and accumulated more Na(+) in the shoots compared with the wild type. OsHKT1;1 was expressed mainly in the phloem of leaf blades and up-regulated in response to salt stress. Using a yeast one-hybrid approach, a novel MYB coiled-coil type transcription factor, OsMYBc, was found to bind to the OsHKT1;1 promoter. In vivo chromatin immunoprecipitation and in vitro electrophoresis mobility shift assays demonstrated that OsMYBc binds to AAANATNC(C/T) fragments within the OsHKT1;1 promoter. Mutation of the OsMYBc-binding nucleotides resulted in a decrease in promoter activity of OsHKT1;1. Knockout of OsMYBc resulted in a reduction in NaCl-induced expression of OsHKT1;1 and salt sensitivity. Taken together, these results suggest that OsHKT1;1 has a role in controlling Na(+) concentration and preventing sodium toxicity in leaf blades and is regulated by the OsMYBc transcription factor.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures
Similar articles
-
A Magnesium Transporter OsMGT1 Plays a Critical Role in Salt Tolerance in Rice.Plant Physiol. 2017 Jul;174(3):1837-1849. doi: 10.1104/pp.17.00532. Epub 2017 May 9. Plant Physiol. 2017. PMID: 28487477 Free PMC article.
-
OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice.Plant J. 2017 Aug;91(4):657-670. doi: 10.1111/tpj.13595. Epub 2017 Jun 13. Plant J. 2017. PMID: 28488420
-
The transcription factor OsMYBc and an E3 ligase regulate expression of a K+ transporter during salt stress.Plant Physiol. 2022 Aug 29;190(1):843-859. doi: 10.1093/plphys/kiac283. Plant Physiol. 2022. PMID: 35695778 Free PMC article.
-
A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K(+)/Na(+) ratio in leaves during salinity stress.Plant Cell Environ. 2010 Apr;33(4):552-65. doi: 10.1111/j.1365-3040.2009.02056.x. Epub 2009 Nov 4. Plant Cell Environ. 2010. PMID: 19895406 Review.
-
Plant HKT Channels: An Updated View on Structure, Function and Gene Regulation.Int J Mol Sci. 2021 Feb 14;22(4):1892. doi: 10.3390/ijms22041892. Int J Mol Sci. 2021. PMID: 33672907 Free PMC article. Review.
Cited by
-
Trehalose alleviates salt tolerance by improving photosynthetic performance and maintaining mineral ion homeostasis in tomato plants.Front Plant Sci. 2022 Aug 12;13:974507. doi: 10.3389/fpls.2022.974507. eCollection 2022. Front Plant Sci. 2022. PMID: 36035709 Free PMC article.
-
Genome-Wide Association Study Reveals Candidate Genes for Root-Related Traits in Rice.Curr Issues Mol Biol. 2022 Sep 21;44(10):4386-4405. doi: 10.3390/cimb44100301. Curr Issues Mol Biol. 2022. PMID: 36286016 Free PMC article.
-
Mechanism of high affinity potassium transporter (HKT) towards improved crop productivity in saline agricultural lands.3 Biotech. 2022 Feb;12(2):51. doi: 10.1007/s13205-021-03092-0. Epub 2022 Jan 27. 3 Biotech. 2022. PMID: 35127306 Free PMC article. Review.
-
Advances in understanding salt tolerance in rice.Theor Appl Genet. 2019 Apr;132(4):851-870. doi: 10.1007/s00122-019-03301-8. Epub 2019 Feb 13. Theor Appl Genet. 2019. PMID: 30759266 Review.
-
Identification of Candidate Genes for Salt Tolerance at the Seedling Stage Using Integrated Genome-Wide Association Study and Transcriptome Analysis in Rice.Plants (Basel). 2023 Mar 21;12(6):1401. doi: 10.3390/plants12061401. Plants (Basel). 2023. PMID: 36987089 Free PMC article.
References
-
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 - PubMed
-
- Apse MP, Blumwald E (2007) Na+ transport in plants. FEBS Lett 581: 2247–2254 - PubMed
-
- Baek D, Jiang J, Chung JS, Wang B, Chen J, Xin Z, Shi H (2011) Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol 52: 149–161 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

