Parasexual Ploidy Reduction Drives Population Heterogeneity Through Random and Transient Aneuploidy in Candida albicans
- PMID: 25991822
- PMCID: PMC4512543
- DOI: 10.1534/genetics.115.178020
Parasexual Ploidy Reduction Drives Population Heterogeneity Through Random and Transient Aneuploidy in Candida albicans
Abstract
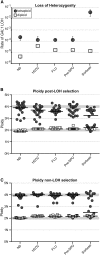
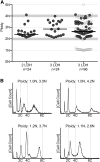
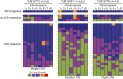
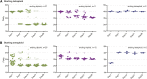
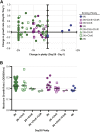
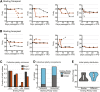
The opportunistic pathogen Candida albicans has a large repertoire of mechanisms to generate genetic and phenotypic diversity despite the lack of meiosis in its life cycle. Its parasexual cycle enables shifts in ploidy, which in turn facilitate recombination, aneuploidy, and homozygosis of whole chromosomes to fuel rapid adaptation. Here we show that the tetraploid state potentiates ploidy variation and drives population heterogeneity. In tetraploids, the rate of losing a single heterozygous marker [loss of heterozygosity (LOH)] is elevated ∼30-fold higher than the rate in diploid cells. Furthermore, isolates recovered after selection for LOH of one, two, or three markers were highly aneuploid, with a broad range of karyotypes including strains with a combination of di-, tri-, and tetrasomic chromosomes. We followed the ploidy trajectories for these tetraploid- and aneuploid-derived isolates, using a combination of flow cytometry and double-digestion restriction-site-associated DNA analyzed with next-generation sequencing. Isolates derived from either tetraploid or aneuploid isolates predominately resolved to a stable euploid state. The majority of isolates reduced to the conventional diploid state; however, stable triploid and tetraploid states were observed in ∼30% of the isolates. Notably, aneuploid isolates were more transient than tetraploid isolates, resolving to a euploid state within a few passages. Furthermore, the likelihood that a particular isolate will resolve to the same ploidy state in replicate evolution experiments is only ∼50%, supporting the idea that the chromosome loss process of the parasexual cycle is random and does not follow trajectories involving specific combinations of chromosomes. Together, our results indicate that tetraploid progenitors can produce populations of progeny cells with a high degree of genomic diversity, from altered ploidy to homozygosis, providing an excellent source of genetic variation upon which selection can act.
Keywords: parasex; ploidy; yeast.
Copyright © 2015 by the Genetics Society of America.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

