Rapid molecular evolution across amniotes of the IIS/TOR network
- PMID: 25991861
- PMCID: PMC4460498
- DOI: 10.1073/pnas.1419659112
Rapid molecular evolution across amniotes of the IIS/TOR network
Abstract
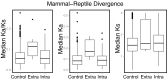
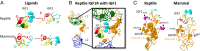
The insulin/insulin-like signaling and target of rapamycin (IIS/TOR) network regulates lifespan and reproduction, as well as metabolic diseases, cancer, and aging. Despite its vital role in health, comparative analyses of IIS/TOR have been limited to invertebrates and mammals. We conducted an extensive evolutionary analysis of the IIS/TOR network across 66 amniotes with 18 newly generated transcriptomes from nonavian reptiles and additional available genomes/transcriptomes. We uncovered rapid and extensive molecular evolution between reptiles (including birds) and mammals: (i) the IIS/TOR network, including the critical nodes insulin receptor substrate (IRS) and phosphatidylinositol 3-kinase (PI3K), exhibit divergent evolutionary rates between reptiles and mammals; (ii) compared with a proxy for the rest of the genome, genes of the IIS/TOR extracellular network exhibit exceptionally fast evolutionary rates; and (iii) signatures of positive selection and coevolution of the extracellular network suggest reptile- and mammal-specific interactions between members of the network. In reptiles, positively selected sites cluster on the binding surfaces of insulin-like growth factor 1 (IGF1), IGF1 receptor (IGF1R), and insulin receptor (INSR); whereas in mammals, positively selected sites clustered on the IGF2 binding surface, suggesting that these hormone-receptor binding affinities are targets of positive selection. Further, contrary to reports that IGF2R binds IGF2 only in marsupial and placental mammals, we found positively selected sites clustered on the hormone binding surface of reptile IGF2R that suggest that IGF2R binds to IGF hormones in diverse taxa and may have evolved in reptiles. These data suggest that key IIS/TOR paralogs have sub- or neofunctionalized between mammals and reptiles and that this network may underlie fundamental life history and physiological differences between these amniote sister clades.
Keywords: insulin growth factor; insulin signaling; molecular evolution; rapamycin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. - PubMed
-
- Kenyon CJ. The genetics of ageing. Nature. 2010;464(7288):504–512. - PubMed
-
- Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: Insights into insulin action. Nat Rev Mol Cell Biol. 2006;7(2):85–96. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

