Atlas of tissue renin-angiotensin-aldosterone system in human: A transcriptomic meta-analysis
- PMID: 25992767
- PMCID: PMC4445654
- DOI: 10.1038/srep10035
Atlas of tissue renin-angiotensin-aldosterone system in human: A transcriptomic meta-analysis
Abstract
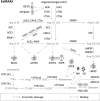
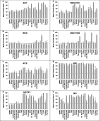
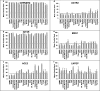
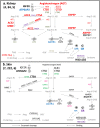
Tissue renin-angiotensin-aldosterone system (RAAS) has attracted much attention because of its physiological and pharmacological implications; however, a clear definition of tissue RAAS is still missing. We aimed to establish a preliminary atlas for the organization of RAAS across 23 different normal human tissues. A set of 37 genes encoding classical and novel RAAS participants including gluco- and mineralo-corticoids were defined as extended RAAS (extRAAS) system. Microarray data sets containing more than 10 normal tissues were downloaded from the GEO database. R software was used to extract expression levels and construct dendrograms of extRAAS genes within each data set. Tissue co-expression modules were then extracted from reproducible gene clusters across data sets. An atlas of the maps of tissue-specific organization of extRAAS was constructed from gene expression and coordination data. Our analysis included 143 data sets containing 4933 samples representing 23 different tissues. Expression data provided an insight on the favored pathways in a given tissue. Gene coordination indicated the existence of tissue-specific modules organized or not around conserved core groups of transcripts. The atlas of tissue-specific organization of extRAAS will help better understand tissue-specific effects of RAAS. This will provide a frame for developing more effective and selective pharmaceuticals targeting extRAAS.
Figures



References
-
- Robertson J. I. S., Nichools M. G. & August P. The renin-angiotensin system . Mosby1993).
-
- Michel J.-B. Médecine cardiovasculaire du système rénine angiotensine. (Ed. Pradel, 1992).
-
- Paul M., Mehr A. P. & Kreutz R. Physiology of Local Renin-Angiotensin Systems. Physiol. Rev. 86, 747–803 (2006). - PubMed
-
- Uehara Y., Miura S., Yahiro E. & Saku K. Non-ACE pathway-induced angiotensin II production. Curr. Pharm. Des. 19, 3054–3059 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

