Model-Free RNA Sequence and Structure Alignment Informed by SHAPE Probing Reveals a Conserved Alternate Secondary Structure for 16S rRNA
- PMID: 25992778
- PMCID: PMC4438973
- DOI: 10.1371/journal.pcbi.1004126
Model-Free RNA Sequence and Structure Alignment Informed by SHAPE Probing Reveals a Conserved Alternate Secondary Structure for 16S rRNA
Abstract
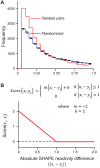
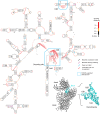
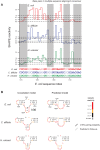
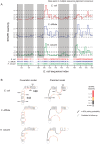
Discovery and characterization of functional RNA structures remains challenging due to deficiencies in de novo secondary structure modeling. Here we describe a dynamic programming approach for model-free sequence comparison that incorporates high-throughput chemical probing data. Based on SHAPE probing data alone, ribosomal RNAs (rRNAs) from three diverse organisms--the eubacteria E. coli and C. difficile and the archeon H. volcanii--could be aligned with accuracies comparable to alignments based on actual sequence identity. When both base sequence identity and chemical probing reactivities were considered together, accuracies improved further. Derived sequence alignments and chemical probing data from protein-free RNAs were then used as pseudo-free energy constraints to model consensus secondary structures for the 16S and 23S rRNAs. There are critical differences between these experimentally-informed models and currently accepted models, including in the functionally important neck and decoding regions of the 16S rRNA. We infer that the 16S rRNA has evolved to undergo large-scale changes in base pairing as part of ribosome function. As high-quality RNA probing data become widely available, structurally-informed sequence alignment will become broadly useful for de novo motif and function discovery.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Gesteland RF, Cech T, Atkins JF. The RNA World. 3rd ed Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2006.
-
- Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216(3): 585–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

