Antibodies to Interleukin-2 Elicit Selective T Cell Subset Potentiation through Distinct Conformational Mechanisms
- PMID: 25992858
- PMCID: PMC4439582
- DOI: 10.1016/j.immuni.2015.04.015
Antibodies to Interleukin-2 Elicit Selective T Cell Subset Potentiation through Distinct Conformational Mechanisms
Abstract
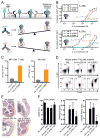
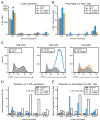
Interleukin-2 (IL-2) is a pleiotropic cytokine that regulates immune cell homeostasis and has been used to treat a range of disorders including cancer and autoimmune disease. IL-2 signals via interleukin-2 receptor-β (IL-2Rβ):IL-2Rγ heterodimers on cells expressing high (regulatory T cells, Treg) or low (effector cells) amounts of IL-2Rα (CD25). When complexed with IL-2, certain anti-cytokine antibodies preferentially stimulate expansion of Treg (JES6-1) or effector (S4B6) cells, offering a strategy for targeted disease therapy. We found that JES6-1 sterically blocked the IL-2:IL-2Rβ and IL-2:IL-2Rγ interactions, but also allosterically lowered the IL-2:IL-2Rα affinity through a "triggered exchange" mechanism favoring IL-2Rα(hi) Treg cells, creating a positive feedback loop for IL-2Rα(hi) cell activation. Conversely, S4B6 sterically blocked the IL-2:IL-2Rα interaction, while also conformationally stabilizing the IL-2:IL-2Rβ interaction, thus stimulating all IL-2-responsive immune cells, particularly IL-2Rβ(hi) effector cells. These insights provide a molecular blueprint for engineering selectively potentiating therapeutic antibodies.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors have no other competing financial interests.
Figures




Comment in
-
IL-2: Change Structure … Change Function.Immunity. 2015 May 19;42(5):779-81. doi: 10.1016/j.immuni.2015.05.002. Immunity. 2015. PMID: 25992850
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
-
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nature biotechnology. 1997;15:553–557. - PubMed
-
- Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006a;311:1924–1927. - PubMed
-
- Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12:180–190. - PubMed
-
- Boyman O, Surh CD, Sprent J. Potential use of IL-2/anti-IL-2 antibody immune complexes for the treatment of cancer and autoimmune disease. Expert Opin Biol Ther. 2006b;6:1323–1331. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

