Asymmetric Action of STAT Transcription Factors Drives Transcriptional Outputs and Cytokine Specificity
- PMID: 25992861
- PMCID: PMC11037422
- DOI: 10.1016/j.immuni.2015.04.014
Asymmetric Action of STAT Transcription Factors Drives Transcriptional Outputs and Cytokine Specificity
Abstract
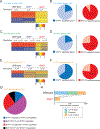
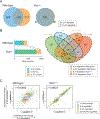
Interleukin-6 (IL-6) and IL-27 signal through a shared receptor subunit and employ the same downstream STAT transcription proteins, but yet are ascribed unique and overlapping functions. To evaluate the specificity and redundancy for these cytokines, we quantified their global transcriptomic changes and determined the relative contributions of STAT1 and STAT3 using genetic models and chromatin immunoprecipitation-sequencing (ChIP-seq) approaches. We found an extensive overlap of the transcriptomes induced by IL-6 and IL-27 and few examples in which the cytokines acted in opposition. Using STAT-deficient cells and T cells from patients with gain-of-function STAT1 mutations, we demonstrated that STAT3 is responsible for the overall transcriptional output driven by both cytokines, whereas STAT1 is the principal driver of specificity. STAT1 cannot compensate in the absence of STAT3 and, in fact, much of STAT1 binding to chromatin is STAT3 dependent. Thus, STAT1 shapes the specific cytokine signature superimposed upon STAT3's action.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures





References
-
- Awasthi A, Carrier Y, Peron JP, Bettelli E, Kamanaka M, Flavell RA, Kuchroo VK, Oukka M, and Weiner HL (2007). A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat Immunol 8, 1380–1389. - PubMed
-
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, and Kuchroo VK (2006). Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

