Clonal Deletion Prunes but Does Not Eliminate Self-Specific αβ CD8(+) T Lymphocytes
- PMID: 25992863
- PMCID: PMC4455602
- DOI: 10.1016/j.immuni.2015.05.001
Clonal Deletion Prunes but Does Not Eliminate Self-Specific αβ CD8(+) T Lymphocytes
Abstract
It has long been thought that clonal deletion efficiently removes almost all self-specific T cells from the peripheral repertoire. We found that self-peptide MHC-specific CD8(+) T cells in the blood of healthy humans were present in frequencies similar to those specific for non-self antigens. For the Y chromosome-encoded SMCY antigen, self-specific T cells exhibited only a 3-fold lower average frequency in males versus females and were anergic with respect to peptide activation, although this inhibition could be overcome by a stronger stimulus. We conclude that clonal deletion prunes but does not eliminate self-specific T cells and suggest that to do so would create holes in the repertoire that pathogens could readily exploit. In support of this hypothesis, we detected T cells specific for all 20 amino acid variants at the p5 position of a hepatitis C virus epitope in a random group of blood donors.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
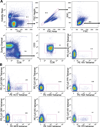
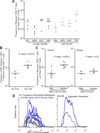
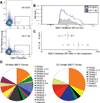


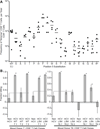
Comment in
-
Thymic selection: to thine own self be true.Immunity. 2015 May 19;42(5):788-9. doi: 10.1016/j.immuni.2015.05.007. Immunity. 2015. PMID: 25992854
References
-
- Alanio C, Lemaitre F, Law HK, Hasan M, Albert ML. Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood. 2010;115:3718–3725. - PubMed
-
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. - PubMed
-
- Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GM, Papagno L, Ogg GS, King A, Lechner F, Spina CA, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8:379–385. - PubMed
-
- Barnea E, Beer I, Patoka R, Ziv T, Kessler O, Tzehoval E, Eisenbach L, Zavazava N, Admon A. Analysis of endogenous peptides bound by soluble MHC class I molecules: a novel approach for identifying tumor-specific antigens. Eur J Immunol. 2002;32:213–222. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

