Structure-Based Alignment and Consensus Secondary Structures for Three HIV-Related RNA Genomes
- PMID: 25992893
- PMCID: PMC4439019
- DOI: 10.1371/journal.pcbi.1004230
Structure-Based Alignment and Consensus Secondary Structures for Three HIV-Related RNA Genomes
Abstract
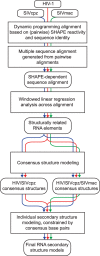
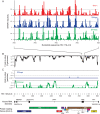
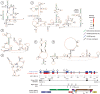
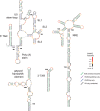
HIV and related primate lentiviruses possess single-stranded RNA genomes. Multiple regions of these genomes participate in critical steps in the viral replication cycle, and the functions of many RNA elements are dependent on the formation of defined structures. The structures of these elements are still not fully understood, and additional functional elements likely exist that have not been identified. In this work, we compared three full-length HIV-related viral genomes: HIV-1NL4-3, SIVcpz, and SIVmac (the latter two strains are progenitors for all HIV-1 and HIV-2 strains, respectively). Model-free RNA structure comparisons were performed using whole-genome structure information experimentally derived from nucleotide-resolution SHAPE reactivities. Consensus secondary structures were constructed for strongly correlated regions by taking into account both SHAPE probing structural data and nucleotide covariation information from structure-based alignments. In these consensus models, all known functional RNA elements were recapitulated with high accuracy. In addition, we identified multiple previously unannotated structural elements in the HIV-1 genome likely to function in translation, splicing and other replication cycle processes; these are compelling targets for future functional analyses. The structure-informed alignment strategy developed here will be broadly useful for efficient RNA motif discovery.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs.PLoS Pathog. 2013;9(4):e1003294. doi: 10.1371/journal.ppat.1003294. Epub 2013 Apr 4. PLoS Pathog. 2013. PMID: 23593004 Free PMC article.
-
High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states.PLoS Biol. 2008 Apr 29;6(4):e96. doi: 10.1371/journal.pbio.0060096. PLoS Biol. 2008. PMID: 18447581 Free PMC article.
-
Mapping the RNA structural landscape of viral genomes.Methods. 2020 Nov 1;183:57-67. doi: 10.1016/j.ymeth.2019.11.001. Epub 2019 Nov 8. Methods. 2020. PMID: 31711930 Free PMC article.
-
Physical and Functional Analysis of Viral RNA Genomes by SHAPE.Annu Rev Virol. 2019 Sep 29;6(1):93-117. doi: 10.1146/annurev-virology-092917-043315. Epub 2019 Jul 23. Annu Rev Virol. 2019. PMID: 31337286 Free PMC article. Review.
-
RNA structure analysis of alphacoronavirus terminal genome regions.Virus Res. 2014 Dec 19;194:76-89. doi: 10.1016/j.virusres.2014.10.001. Epub 2014 Oct 13. Virus Res. 2014. PMID: 25307890 Free PMC article. Review.
Cited by
-
RNA secondary structure packages evaluated and improved by high-throughput experiments.Nat Methods. 2022 Oct;19(10):1234-1242. doi: 10.1038/s41592-022-01605-0. Epub 2022 Oct 3. Nat Methods. 2022. PMID: 36192461 Free PMC article.
-
Guidelines for SHAPE Reagent Choice and Detection Strategy for RNA Structure Probing Studies.Biochemistry. 2019 Jun 11;58(23):2655-2664. doi: 10.1021/acs.biochem.8b01218. Epub 2019 May 30. Biochemistry. 2019. PMID: 31117385 Free PMC article.
-
The Effect of RNA Substitution Models on Viroid and RNA Virus Phylogenies.Genome Biol Evol. 2018 Feb 1;10(2):657-666. doi: 10.1093/gbe/evx273. Genome Biol Evol. 2018. PMID: 29325030 Free PMC article.
-
Structure-first identification of RNA elements that regulate dengue virus genome architecture and replication.Proc Natl Acad Sci U S A. 2023 Apr 11;120(15):e2217053120. doi: 10.1073/pnas.2217053120. Epub 2023 Apr 3. Proc Natl Acad Sci U S A. 2023. PMID: 37011200 Free PMC article.
-
Bridging the gap between in vitro and in vivo RNA folding.Q Rev Biophys. 2016 Jan;49:e10. doi: 10.1017/S003358351600007X. Epub 2016 Jun 24. Q Rev Biophys. 2016. PMID: 27658939 Free PMC article.
References
-
- Gesteland RF, Cech T, Atkins JF. The RNA World. 3rd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2006.
-
- Coffin JM, Hughes SH, Varmus HE. Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Cold Spring Harbor (NY)1997.
-
- Muesing MA, Smith DH, Capon DJ. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48(4): 691–701. - PubMed
-
- Olsen HS, Nelbock P, Cochrane AW, Rosen CA. Secondary structure is the major determinant for interaction of HIV rev protein with RNA. Science. 1990;247(4944): 845–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

