Convergent evolution at the gametophytic self-incompatibility system in Malus and Prunus
- PMID: 25993016
- PMCID: PMC4438004
- DOI: 10.1371/journal.pone.0126138
Convergent evolution at the gametophytic self-incompatibility system in Malus and Prunus
Abstract
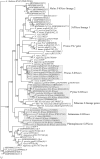
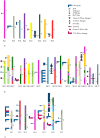
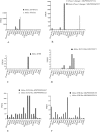
S-RNase-based gametophytic self-incompatibility (GSI) has evolved once before the split of the Asteridae and Rosidae. This conclusion is based on the phylogenetic history of the S-RNase that determines pistil specificity. In Rosaceae, molecular characterizations of Prunus species, and species from the tribe Pyreae (i.e., Malus, Pyrus, Sorbus) revealed different numbers of genes determining S-pollen specificity. In Prunus only one pistil and pollen gene determine GSI, while in Pyreae there is one pistil but multiple pollen genes, implying different specificity recognition mechanisms. It is thus conceivable that within Rosaceae the genes involved in GSI in the two lineages are not orthologous but possibly paralogous. To address this hypothesis we characterised the S-RNase lineage and S-pollen lineage genes present in the genomes of five Rosaceae species from three genera: M. × domestica (apple, self-incompatible (SI); tribe Pyreae), P. persica (peach, self-compatible (SC); Amygdaleae), P. mume (mei, SI; Amygdaleae), Fragaria vesca (strawberry, SC; Potentilleae), and F. nipponica (mori-ichigo, SI; Potentilleae). Phylogenetic analyses revealed that the Malus and Prunus S-RNase and S-pollen genes belong to distinct gene lineages, and that only Prunus S-RNase and SFB-lineage genes are present in Fragaria. Thus, S-RNase based GSI system of Malus evolved independently from the ancestral system of Rosaceae. Using expression patterns based on RNA-seq data, the ancestral S-RNase lineage gene is inferred to be expressed in pistils only, while the ancestral S-pollen lineage gene is inferred to be expressed in tissues other than pollen.
Conflict of interest statement
Figures


References
-
- De Nettancourt D (1977) Incompatibility in angiosperms: Springer-Verlag, Berlin.
-
- Igic B, Lande R, Kohn JR (2008) Loss of self‐incompatibility and its evolutionary consequences. International Journal of Plant Sciences 169: 93–104.
-
- Roalson EH, McCubbin AG (2003) S-RNases and sexual incompatibility: structure, functions, and evolutionary perspectives. Molecular Phylogenetics and Evolution 29: 490–506. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

