MALDI-Mass Spectrometric Imaging Revealing Hypoxia-Driven Lipids and Proteins in a Breast Tumor Model
- PMID: 25993305
- PMCID: PMC4820759
- DOI: 10.1021/ac504503x
MALDI-Mass Spectrometric Imaging Revealing Hypoxia-Driven Lipids and Proteins in a Breast Tumor Model
Abstract
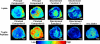
Hypoxic areas are a common feature of rapidly growing malignant tumors and their metastases and are typically spatially heterogeneous. Hypoxia has a strong impact on tumor cell biology and contributes to tumor progression in multiple ways. To date, only a few molecular key players in tumor hypoxia, such as hypoxia-inducible factor-1 (HIF-1), have been discovered. The distribution of biomolecules is frequently heterogeneous in the tumor volume and may be driven by hypoxia and HIF-1α. Understanding the spatially heterogeneous hypoxic response of tumors is critical. Mass spectrometric imaging (MSI) provides a unique way of imaging biomolecular distributions in tissue sections with high spectral and spatial resolution. In this paper, breast tumor xenografts grown from MDA-MB-231-HRE-tdTomato cells, with a red fluorescent tdTomato protein construct under the control of a hypoxia response element (HRE)-containing promoter driven by HIF-1α, were used to detect the spatial distribution of hypoxic regions. We elucidated the 3D spatial relationship between hypoxic regions and the localization of lipids and proteins by using principal component analysis-linear discriminant analysis (PCA-LDA) on 3D rendered MSI volume data from MDA-MB-231-HRE-tdTomato breast tumor xenografts. In this study, we identified hypoxia-regulated proteins active in several distinct pathways such as glucose metabolism, regulation of actin cytoskeleton, protein folding, translation/ribosome, splicesome, the PI3K-Akt signaling pathway, hemoglobin chaperone, protein processing in endoplasmic reticulum, detoxification of reactive oxygen species, aurora B signaling/apoptotic execution phase, the RAS signaling pathway, the FAS signaling pathway/caspase cascade in apoptosis, and telomere stress induced senescence. In parallel, we also identified colocalization of hypoxic regions and various lipid species such as PC(16:0/18:0), PC(16:0/18:1), PC(16:0/18:2), PC(16:1/18:4), PC(18:0/18:1), and PC(18:1/18:1), among others. Our findings shed light on the biomolecular composition of hypoxic tumor regions, which may be responsible for a given tumor's resistance to radiation or chemotherapy.
Figures



References
-
- Hockel M, Vaupel PJ. Natl. Cancer Inst. 2001;93:266–276. - PubMed
-
- Tatum JL, Kelloff GJ, Gillies RJ, Arbeit JM, Brown JM, Chao KS, Chapman JD, Eckelman WC, Fyles AW, Giaccia AJ, Hill RP, Koch CJ, Krishna MC, Krohn KA, Lewis JS, Mason RP, Melillo G, Padhani AR, Powis G, Rajendran JG, Reba R, Robinson SP, Semenza GL, Swartz HM, Vaupel P, Yang D, Croft B, Hoffman J, Liu G, Stone H, Sullivan D. Int. J. Radiat. Biol. 2006;82:699–757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

