Two Novel Motifs of Watermelon Silver Mottle Virus NSs Protein Are Responsible for RNA Silencing Suppression and Pathogenicity
- PMID: 25993336
- PMCID: PMC4439075
- DOI: 10.1371/journal.pone.0126161
Two Novel Motifs of Watermelon Silver Mottle Virus NSs Protein Are Responsible for RNA Silencing Suppression and Pathogenicity
Abstract
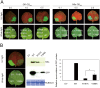
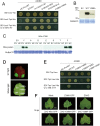
The NSs protein of Watermelon silver mottle virus (WSMoV) is the RNA silencing suppressor and pathogenicity determinant. In this study, serial deletion and point-mutation mutagenesis of conserved regions (CR) of NSs protein were performed, and the silencing suppression function was analyzed through agroinfiltration in Nicotiana benthamiana plants. We found two amino acid (aa) residues, H113 and Y398, are novel functional residues for RNA silencing suppression. Our further analyses demonstrated that H113 at the common epitope (CE) ((109)KFTMHNQ(117)), which is highly conserved in Asia type tospoviruses, and the benzene ring of Y398 at the C-terminal β-sheet motif ((397)IYFL(400)) affect NSs mRNA stability and protein stability, respectively, and are thus critical for NSs RNA silencing suppression. Additionally, protein expression of other six deleted (ΔCR1-ΔCR6) and five point-mutated (Y15A, Y27A, G180A, R181A and R212A) mutants were hampered and their silencing suppression ability was abolished. The accumulation of the mutant mRNAs and proteins, except Y398A, could be rescued or enhanced by co-infiltration with potyviral suppressor HC-Pro. When assayed with the attenuated Zucchini yellow mosaic virus vector in squash plants, the recombinants carrying individual seven point-mutated NSs proteins displayed symptoms much milder than the recombinant carrying the wild type NSs protein, suggesting that these aa residues also affect viral pathogenicity by suppressing the host silencing mechanism.
Conflict of interest statement
Figures




References
-
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000; 101: 533–542. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

