Oligonucleotide Probes for ND-FISH Analysis to Identify Rye and Wheat Chromosomes
- PMID: 25994088
- PMCID: PMC4440213
- DOI: 10.1038/srep10552
Oligonucleotide Probes for ND-FISH Analysis to Identify Rye and Wheat Chromosomes
Abstract
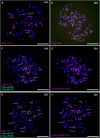
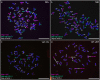
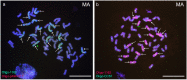
Genomic in situ hybridization (GISH) has been widely used to detect rye (Secale cereale L.) chromosomes in wheat (Triticum aestivum L.) introgression lines. The routine procedure of GISH using genomic DNA of rye as a probe is time-consuming and labor-intensive because of the preparation and labeling of genomic DNA of rye and denaturing of chromosomes and probes. In this study, new oligonucleotide probes Oligo-1162, Oligo-pSc200 and Oligo-pSc250 were developed. The three new probes can be used for non-denaturing fluorescence in situ hybridization (ND-FISH) assays and replace genomic DNA of rye as a probe to discriminate rye chromosomes in wheat backgrounds. In addition, previously developed oligonucleotide probes Oligo-pSc119.2-1, Oligo-pSc119.2-2, Oligo-pTa535-1, Oligo-pTa535-2, Oligo-pTa71-2, Oligo-pAWRC.1 and Oligo-CCS1 can also be used for ND-FISH of wheat and rye. These probes have provided an easier, faster and more cost-effective method for the FISH analysis of wheat and hybrids derived from wheat × rye.
Figures

Similar articles
-
Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis.J Appl Genet. 2014 Aug;55(3):313-8. doi: 10.1007/s13353-014-0215-z. Epub 2014 Apr 30. J Appl Genet. 2014. PMID: 24782110
-
Oligonucleotides and ND-FISH Displaying Different Arrangements of Tandem Repeats and Identification of Dasypyrum villosum Chromosomes in Wheat Backgrounds.Molecules. 2017 Jun 14;22(6):973. doi: 10.3390/molecules22060973. Molecules. 2017. PMID: 28613230 Free PMC article.
-
New types of wheat chromosomal structural variations in derivatives of wheat-rye hybrids.PLoS One. 2014 Oct 10;9(10):e110282. doi: 10.1371/journal.pone.0110282. eCollection 2014. PLoS One. 2014. PMID: 25302962 Free PMC article.
-
Single Copy Oligonucleotide Fluorescence In Situ Hybridization Probe Design Platforms: Development, Application and Evaluation.Int J Mol Sci. 2021 Jul 1;22(13):7124. doi: 10.3390/ijms22137124. Int J Mol Sci. 2021. PMID: 34281175 Free PMC article. Review.
-
Fluorescence in situ hybridization in plants: recent developments and future applications.Chromosome Res. 2019 Sep;27(3):153-165. doi: 10.1007/s10577-019-09607-z. Epub 2019 Mar 9. Chromosome Res. 2019. PMID: 30852707 Review.
Cited by
-
Variations of subtelomeric tandem repeats and rDNA on chromosome 1RS arms in the genus Secale and 1BL.1RS translocations.BMC Plant Biol. 2022 Apr 25;22(1):212. doi: 10.1186/s12870-022-03598-6. BMC Plant Biol. 2022. PMID: 35468732 Free PMC article.
-
Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot.Theor Appl Genet. 2019 Jan;132(1):257-272. doi: 10.1007/s00122-018-3214-3. Epub 2018 Oct 29. Theor Appl Genet. 2019. PMID: 30374527
-
Diversified Chromosome Rearrangements Detected in a Wheat‒Dasypyrum breviaristatum Substitution Line Induced by Gamma-Ray Irradiation.Plants (Basel). 2019 Jun 14;8(6):175. doi: 10.3390/plants8060175. Plants (Basel). 2019. PMID: 31207944 Free PMC article.
-
Stable minichromosome and functional neocentromere derived from rye 7R chromosome arm.BMC Plant Biol. 2024 Dec 18;24(1):1185. doi: 10.1186/s12870-024-05918-4. BMC Plant Biol. 2024. PMID: 39695363 Free PMC article.
-
Fluorescence in situ Hybridization Analysis of Oligonucleotide 5S Ribosomal DNA, 45S Ribosomal DNA, and (TTTAGGG)3 Locations in Gloriosa superba L.Cytogenet Genome Res. 2024;164(5-6):276-283. doi: 10.1159/000541706. Epub 2024 Oct 12. Cytogenet Genome Res. 2024. PMID: 39396512 Free PMC article.
References
-
- Friebe B., Jiang J., Raupp W. J., McIntosh R. A. & Gill B. S. Characterization of wheat-alien translocation conferring resistance to diseases and pests: current status. Euphytica 91, 59–87 (1996).
-
- Lukaszewski A. J., Porter D. R., Baker C. A., Rybka K. & Lapinski B. Attempts to transfer Russian wheat aphid resistance from a rye chromosome in Russian triticales to wheat. Crop Sci. 41, 1743–1749 (2001).
-
- Kim W., Johnson J. W., Baenziger P. S., Lukaszewski A. J. & Gaines C. S. Agronomic effect of wheat-rye translocation carrying rye chromatin (1R) from different sources. Crop Sci. 44, 1254–1258 (2004).
-
- Camacho M. V., Matos M., González C., Pérez-Flores V. & Pernaute B. Secale cereale inter-microsatellites (SCIMs): chromosomal location and genetic inheritance. Genetica 123, 303–311 (2005). - PubMed
-
- An D. G. et al. Molecular cytogenetic characterization of a new wheat-rye 4R chromosome translocation line resistant to powdery mildew. Chromosome Res. 21, 419–432 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

