Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity
- PMID: 25994611
- PMCID: PMC5965381
- DOI: 10.1093/femsre/fuv023
Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity
Abstract
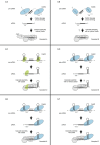
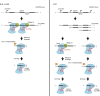
CRISPR-Cas is an RNA-mediated adaptive immune system that defends bacteria and archaea against mobile genetic elements. Short mature CRISPR RNAs (crRNAs) are key elements in the interference step of the immune pathway. A CRISPR array composed of a series of repeats interspaced by spacer sequences acquired from invading mobile genomes is transcribed as a precursor crRNA (pre-crRNA) molecule. This pre-crRNA undergoes one or two maturation steps to generate the mature crRNAs that guide CRISPR-associated (Cas) protein(s) to cognate invading genomes for their destruction. Different types of CRISPR-Cas systems have evolved distinct crRNA biogenesis pathways that implicate highly sophisticated processing mechanisms. In Types I and III CRISPR-Cas systems, a specific endoribonuclease of the Cas6 family, either standalone or in a complex with other Cas proteins, cleaves the pre-crRNA within the repeat regions. In Type II systems, the trans-acting small RNA (tracrRNA) base pairs with each repeat of the pre-crRNA to form a dual-RNA that is cleaved by the housekeeping RNase III in the presence of the protein Cas9. In this review, we present a detailed comparative analysis of pre-crRNA recognition and cleavage mechanisms involved in the biogenesis of guide crRNAs in the three CRISPR-Cas types.
Keywords: Cas5d; Cas6; Cas9; RNase III; crRNA biogenesis; tracrRNA.
© FEMS 2015.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

