Quantitative detection of DNMT3A R882H mutation in acute myeloid leukemia
- PMID: 25994761
- PMCID: PMC4443651
- DOI: 10.1186/s13046-015-0173-2
Quantitative detection of DNMT3A R882H mutation in acute myeloid leukemia
Abstract
Background: DNMT3A mutations represent one of the most frequent gene alterations detectable in acute myeloid leukemia (AML) with normal karyotype. Although various recurrent somatic mutations of DNMT3A have been described, the most common mutation is located at R882 in the methyltransferase domain of the gene. Because of their prognostic significance and high stability during disease evolution, DNMT3A mutations might represent highly informative biomarkers for prognosis and outcome of disease.
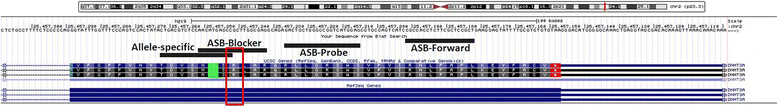
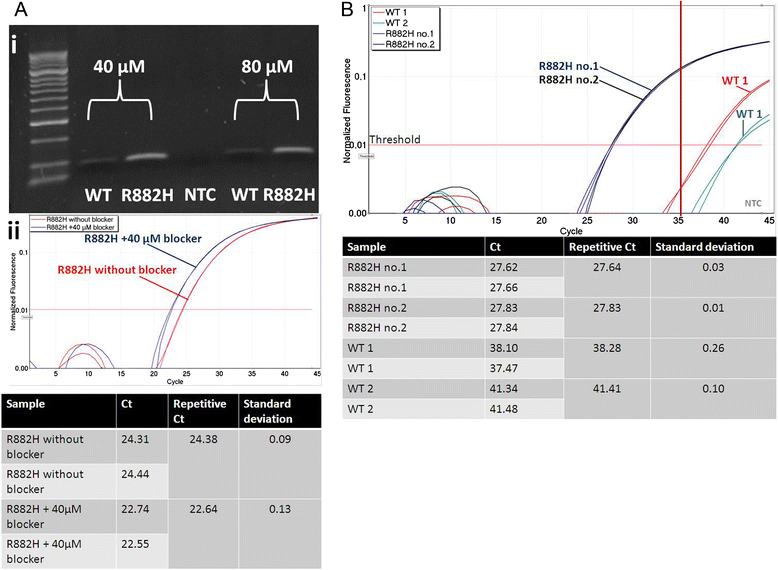
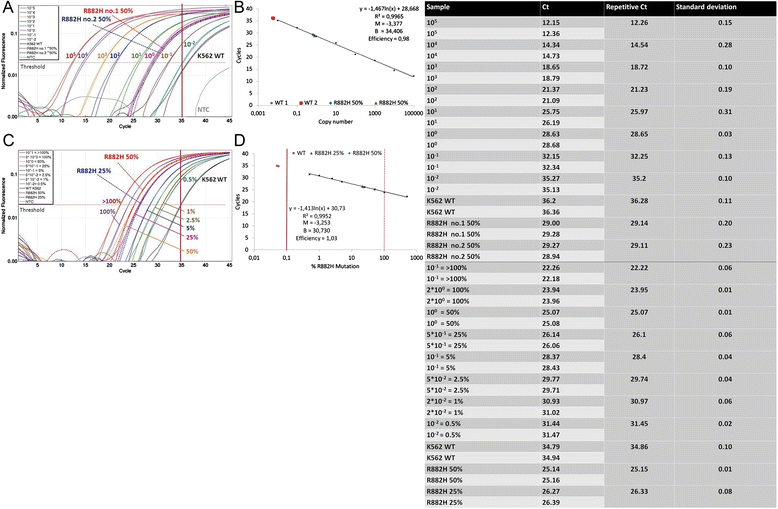
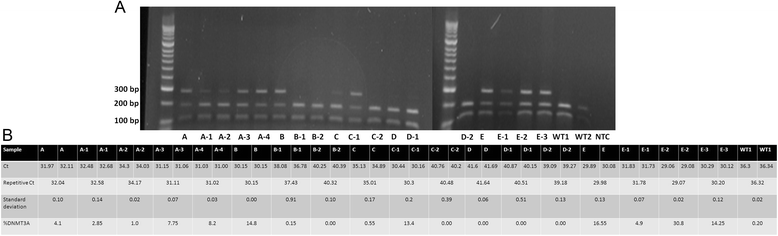
Methods: We describe an allele-specific PCR with a Blocking reagent for the quantitative detection of DNMT3A R882H mutation providing the possibility to analyze the quantitative amount of mutation during the course of disease. Next, we analyzed 62 follow-up samples from 6 AML patients after therapy and allogeneic stem cell transplantation (alloSCT).
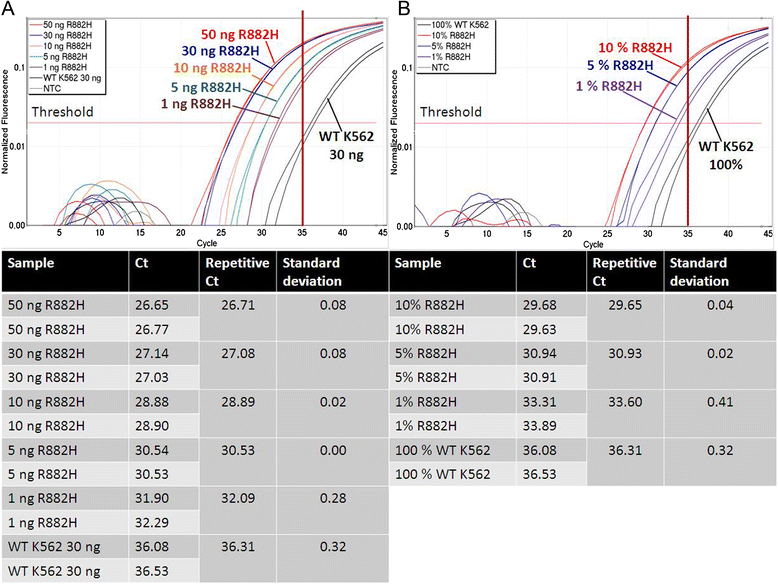
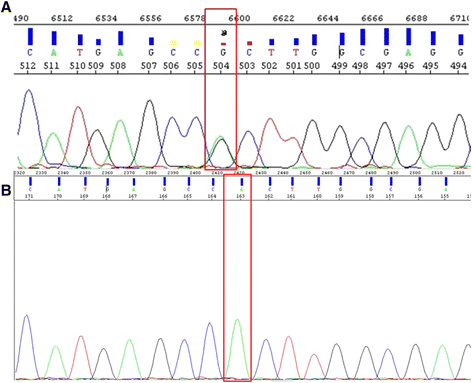
Results: We developed an ASB-PCR assay for quantitative analysis of R882H DNMT3A mutation. After optimization of blocker concentration, a R882H-positive plasmid was constructed to enhance the accuracy of the sensitivity of quantitative detection. The assay displayed a high efficiency and sensitivity up to 10(-3). The reproducibility of assay analyzed using follow-up samples showed the standard deviation less than 3.1 %. This assay displayed a complete concordance with sequencing and endonuclease restriction analysis. We have found persistence of DNMT3A R882H mutations in complete remission (CR) after standard cytoreduction therapy that could be indicating presence of DNMT3A mutation in early pre-leukemic stem cells that resist chemotherapy. The loss of correlation between NPM1 and DNMT3A in CR could be associated with evolution of pre-leukemic and leukemic clones. In patients with CR with complete donor chimerism after alloSCT, we have found no DNMT3A R882H. In relapsed patients, all samples showed an increasing of both NPM1 and DNMT3A mutated alleles. This suggests at least in part the presence of NPM1 and DNMT3A mutations in the same cell clone.
Conclusion: We developed a rapid and reliable method for quantitative detection of DNMT3A R882H mutations in AML patients. Quantitative detection of DNMT3A R882H mutations at different time points of AML disease enables screening of follow-up samples. This could provide additional information about the role of DNMT3A mutations in development and progression of AML.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

