InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams
- PMID: 25994840
- PMCID: PMC4455604
- DOI: 10.1186/s12859-015-0611-3
InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams
Abstract
Background: Set comparisons permeate a large number of data analysis workflows, in particular workflows in biological sciences. Venn diagrams are frequently employed for such analysis but current tools are limited.
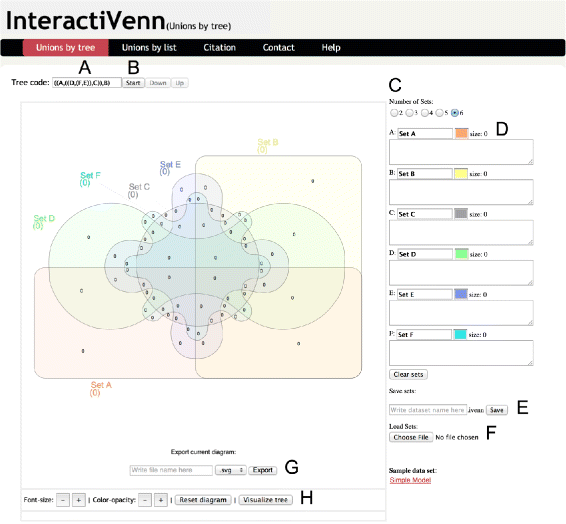
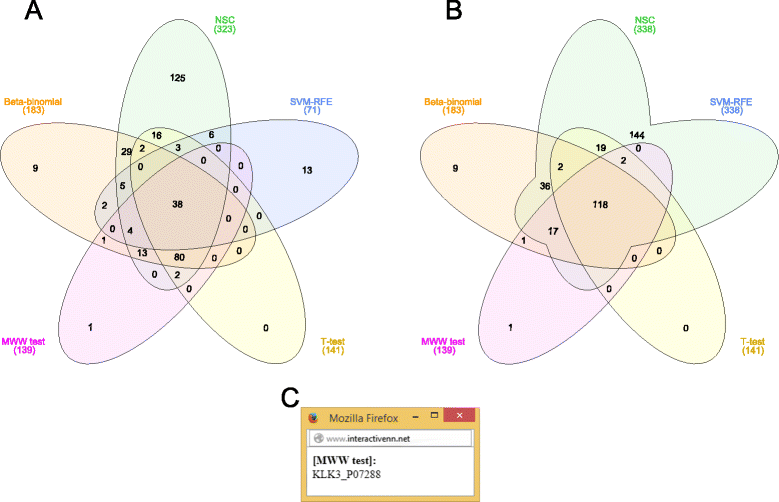
Results: We have developed InteractiVenn, a more flexible tool for interacting with Venn diagrams including up to six sets. It offers a clean interface for Venn diagram construction and enables analysis of set unions while preserving the shape of the diagram. Set unions are useful to reveal differences and similarities among sets and may be guided in our tool by a tree or by a list of set unions. The tool also allows obtaining subsets' elements, saving and loading sets for further analyses, and exporting the diagram in vector and image formats. InteractiVenn has been used to analyze two biological datasets, but it may serve set analysis in a broad range of domains.
Conclusions: InteractiVenn allows set unions in Venn diagrams to be explored thoroughly, by consequence extending the ability to analyze combinations of sets with additional observations, yielded by novel interactions between joined sets. InteractiVenn is freely available online at: www.interactivenn.net .
Figures


References
-
- Ruskey F, Weston M. A survey of Venn diagrams. Electron J Comb. 1997; 4.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

