Characterization of a xylanase-producing Cellvibrio mixtus strain J3-8 and its genome analysis
- PMID: 25994900
- PMCID: PMC4440207
- DOI: 10.1038/srep10521
Characterization of a xylanase-producing Cellvibrio mixtus strain J3-8 and its genome analysis
Abstract
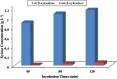
Cellvibrio mixtus strain J3-8 is a gram-negative, xylanase-producing aerobic soil bacterium isolated from giant snails in Singapore. It is able to produce up to 10.1 U ml(-1) of xylanase, which is comparable to xylanase production from known bacterial and fungal strains. Genome sequence analysis of strain J3-8 reveals that the assembled draft genome contains 5,171,890 bp with a G + C content of 46.66%, while open reading frame (ORF) annotations indicate a high density of genes encoding glycoside hydrolase (GH) families involved in (hemi)cellulose hydrolysis. On the basis of 15 identified putative xylanolytic genes, one metabolic pathway in strain J3-8 is constructed for utilization of xylan. In addition, a 1,083 bp xylanase gene from strain J3-8 represents a new member of GH11 family. This gene is verified to be novel via phylogenetic analysis. To utilize this novel gene for hydrolysis of xylan to xylose, it is expressed in recombinant E. coli and characterized for its hydrolytic activity. This study shows that strain J3-8 is a potential candidate for hydrolysis of lignocellulosic materials.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

