Worldwide Population Structure, Long-Term Demography, and Local Adaptation of Helicobacter pylori
- PMID: 25995212
- PMCID: PMC4512554
- DOI: 10.1534/genetics.115.176404
Worldwide Population Structure, Long-Term Demography, and Local Adaptation of Helicobacter pylori
Abstract
Helicobacter pylori is an important human pathogen associated with serious gastric diseases. Owing to its medical importance and close relationship with its human host, understanding genomic patterns of global and local adaptation in H. pylori may be of particular significance for both clinical and evolutionary studies. Here we present the first such whole genome analysis of 60 globally distributed strains, from which we inferred worldwide population structure and demographic history and shed light on interesting global and local events of positive selection, with particular emphasis on the evolution of San-associated lineages. Our results indicate a more ancient origin for the association of humans and H. pylori than previously thought. We identify several important perspectives for future clinical research on candidate selected regions that include both previously characterized genes (e.g., transcription elongation factor NusA and tumor necrosis factor alpha-inducing protein Tipα) and hitherto unknown functional genes.
Keywords: adaptation; human pathogens; neutral evolution.
Copyright © 2015 by the Genetics Society of America.
Figures
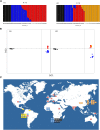
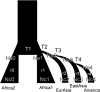

References
-
- Alachiotis N., Stamatakis A., Pavlidis P., 2012. OmegaPlus: a scalable tool for rapid detection of selective sweeps in whole-genome datasets. Bioinformatics 28: 2274–2275. - PubMed
-
- Allaker R. P., Young K. A., Hardie J. M., Domizio P., Meadows N. J., 2002. Prevalence of helicobacter pylori at oral and gastrointestinal sites in children: evidence for possible oral-to-oral transmission. J. Med. Microbiol. 51: 312–317. - PubMed
-
- Argent R. H., Hale J. L., El-Omar E. M., Atherton J. C., 2008. Differences in Helicobacter pylori CagA tyrosine phosphorylation motif patterns between western and East Asian strains, and influences on interleukin-8 secretion. J. Med. Microbiol. 57: 1062–1067. - PubMed
-
- Basso D., Zambon C.-F., Letley D. P., Stranges A., Marchet A., et al. , 2008. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology 135: 91–99. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

