Caribbean and La Réunion Chikungunya Virus Isolates Differ in Their Capacity To Induce Proinflammatory Th1 and NK Cell Responses and Acute Joint Pathology
- PMID: 25995257
- PMCID: PMC4505608
- DOI: 10.1128/JVI.00909-15
Caribbean and La Réunion Chikungunya Virus Isolates Differ in Their Capacity To Induce Proinflammatory Th1 and NK Cell Responses and Acute Joint Pathology
Abstract
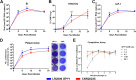
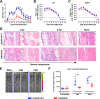
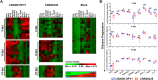
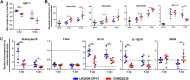
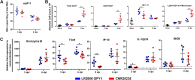
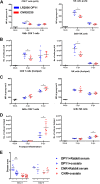
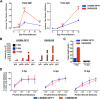
Chikungunya virus (CHIKV) is a mosquito-borne arthralgic alphavirus that has garnered international attention as an important emerging pathogen since 2005. More recently, it invaded the Caribbean islands and the Western Hemisphere. Intriguingly, the current CHIKV outbreak in the Caribbean is caused by the Asian CHIKV genotype, which differs from the La Réunion LR2006 OPY1 isolate belonging to the Indian Ocean lineage. Here, we adopted a systematic and comparative approach against LR2006 OPY1 to characterize the pathogenicity of the Caribbean CNR20235 isolate and consequential host immune responses in mice. Ex vivo infection using primary mouse tail fibroblasts revealed a weaker replication efficiency by CNR20235 isolate. In the CHIKV mouse model, CNR20235 infection induced an enervated joint pathology characterized by moderate edema and swelling, independent of mononuclear cell infiltration. Based on systemic cytokine analysis, localized immunophenotyping, and gene expression profiles in the popliteal lymph node and inflamed joints, two pathogenic phases were defined for CHIKV infection: early acute (2 to 3 days postinfection [dpi]) and late acute (6 to 8 dpi). Reduced joint pathology during early acute phase of CNR20235 infection was associated with a weaker proinflammatory Th1 response and natural killer (NK) cell activity. The pathological role of NK cells was further demonstrated as depletion of NK cells reduced joint pathology in LR2006 OPY1. Taken together, this study provides evidence that the Caribbean CNR20235 isolate has an enfeebled replication and induces a less pathogenic response in the mammalian host.
Importance: The introduction of CHIKV in the Americas has heightened the risk of large-scale outbreaks due to the close proximity between the United States and the Caribbean. The immunopathogenicity of the circulating Caribbean CHIKV isolate was explored, where it was demonstrated to exhibit reduced infectivity resulting in a weakened joint pathology. Analysis of serum cytokine levels, localized immunophenotyping, and gene expression profiles in the organs revealed that a limited Th1 response and reduced NK cells activity could underlie the reduced pathology in the host. Interestingly, higher asymptomatic infections were observed in the Caribbean compared to the La Réunion outbreaks in 2005 and 2006. This is the first study that showed an association between key proinflammatory factors and pathology-mediating leukocytes with a less severe pathological outcome in Caribbean CHIKV infection. Given the limited information regarding the sequela of Caribbean CHIKV infection, our study is timely and will aid the understanding of this increasingly important disease.
Copyright © 2015, Teo et al.
Figures

References
-
- Sergon K, Njuguna C, Kalani R, Ofula V, Onyango C, Konongoi LS, Bedno S, Burke H, Dumilla AM, Konde J, Njenga MK, Sang R, Breiman RF. 2008. Seroprevalence of Chikungunya virus (CHIKV) infection on Lamu Island, Kenya, October 2004. Am J Trop Med Hyg 78:333–337. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

