Conservation of G-Protein Epitopes in Respiratory Syncytial Virus (Group A) Despite Broad Genetic Diversity: Is Antibody Selection Involved in Virus Evolution?
- PMID: 25995258
- PMCID: PMC4505632
- DOI: 10.1128/JVI.00467-15
Conservation of G-Protein Epitopes in Respiratory Syncytial Virus (Group A) Despite Broad Genetic Diversity: Is Antibody Selection Involved in Virus Evolution?
Abstract
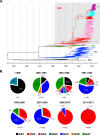
Worldwide G-glycoprotein phylogeny of human respiratory syncytial virus (hRSV) group A sequences revealed diversification in major clades and genotypes over more than 50 years of recorded history. Multiple genotypes cocirculated during prolonged periods of time, but recent dominance of the GA2 genotype was noticed in several studies, and it is highlighted here with sequences from viruses circulating recently in Spain and Panama. Reactivity of group A viruses with monoclonal antibodies (MAbs) that recognize strain-variable epitopes of the G glycoprotein failed to correlate genotype diversification with antibody reactivity. Additionally, no clear correlation was found between changes in strain-variable epitopes and predicted sites of positive selection, despite both traits being associated with the C-terminal third of the G glycoprotein. Hence, our data do not lend support to the proposed antibody-driven selection of variants as a major determinant of hRSV evolution. Other alternative mechanisms are considered to account for the high degree of hRSV G-protein variability.
Importance: An unusual characteristic of the G glycoprotein of human respiratory syncytial virus (hRSV) is the accumulation of nonsynonymous (N) changes at higher rates than synonymous (S) changes, reaching dN/dS values at certain sites predictive of positive selection. Since these sites cluster preferentially in the C-terminal third of the G protein, like certain epitopes recognized by murine antibodies, it was proposed that immune (antibody) selection might be driving the apparent positive selection, analogous to the antigenic drift observed in the influenza virus hemagglutinin (HA). However, careful antigenic and genetic comparison of the G glycoprotein does not provide evidence of antigenic drift in the G molecule, in agreement with recently published data which did not indicate antigenic drift in the G protein with human sera. Alternative explanations to the immune-driven selection hypothesis are offered to account for the high level of G-protein genetic diversity highlighted in this study.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi:10.1016/S0140-6736(10)60206-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

