Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1
- PMID: 25995365
- PMCID: PMC4466714
- DOI: 10.1073/pnas.1501327112
Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1
Abstract
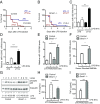
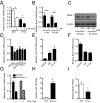
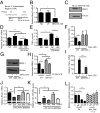
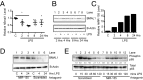
The response to an innate immune challenge is conditioned by the time of day, but the molecular basis for this remains unclear. In myeloid cells, there is a temporal regulation to induction by lipopolysaccharide (LPS) of the proinflammatory microRNA miR-155 that correlates inversely with levels of BMAL1. BMAL1 in the myeloid lineage inhibits activation of NF-κB and miR-155 induction and protects mice from LPS-induced sepsis. Bmal1 has two miR-155-binding sites in its 3'-UTR, and, in response to LPS, miR-155 binds to these two target sites, leading to suppression of Bmal1 mRNA and protein in mice and humans. miR-155 deletion perturbs circadian function, gives rise to a shorter circadian day, and ablates the circadian effect on cytokine responses to LPS. Thus, the molecular clock controls miR-155 induction that can repress BMAL1 directly. This leads to an innate immune response that is variably responsive to challenges across the circadian day.
Keywords: Bmal1; circadian clock; inflammation; miR-155; sepsis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Yang G, et al. Knitting up the raveled sleave of care. Sci Transl Med. 2013;5(212):212rv213. - PubMed
-
- Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LA. Circadian clock proteins and immunity. Immunity. 2014;40(2):178–186. - PubMed
-
- Halberg F, Johnson EA, Brown BW, Bittner JJ. Susceptibility rhythm to E. coli endotoxin and bioassay. Proc Soc Exp Biol Med. 1960;103:142–144. - PubMed
-
- Shackelford PG, Feigin RD. Periodicity of susceptibility to pneumococcal infection: Influence of light and adrenocortical secretions. Science. 1973;182(4109):285–287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

