A Novel Mechanism for Binding of Galactose-terminated Glycans by the C-type Carbohydrate Recognition Domain in Blood Dendritic Cell Antigen 2
- PMID: 25995448
- PMCID: PMC4505424
- DOI: 10.1074/jbc.M115.660613
A Novel Mechanism for Binding of Galactose-terminated Glycans by the C-type Carbohydrate Recognition Domain in Blood Dendritic Cell Antigen 2
Abstract
Blood dendritic cell antigen 2 (BDCA-2; also designated CLEC4C or CD303) is uniquely expressed on plasmacytoid dendritic cells. Stimulation of BDCA-2 with antibodies leads to an anti-inflammatory response in these cells, but the natural ligands for the receptor are not known. The C-type carbohydrate recognition domain in the extracellular portion of BDCA-2 contains a signature motif typical of C-type animal lectins that bind mannose, glucose, or GlcNAc, yet it has been reported that BDCA-2 binds selectively to galactose-terminated, biantennary N-linked glycans. A combination of glycan array analysis and binding competition studies with monosaccharides and natural and synthetic oligosaccharides have been used to define the binding epitope for BDCA-2 as the trisaccharide Galβ1-3/4GlcNAcβ1-2Man. X-ray crystallography and mutagenesis studies show that mannose is ligated to the conserved Ca(2+) in the primary binding site that is characteristic of C-type carbohydrate recognition domains, and the GlcNAc and galactose residues make additional interactions in a wide, shallow groove adjacent to the primary binding site. As predicted from these studies, BDCA-2 binds to IgG, which bears galactose-terminated glycans that are not commonly found attached to other serum glycoproteins. Thus, BDCA-2 has the potential to serve as a previously unrecognized immunoglobulin Fc receptor.
Keywords: CD303; CLEC4C; carbohydrate-binding protein; crystal structure; glycobiology; glycoprotein; lectin.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




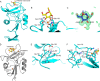
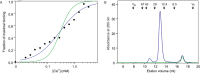


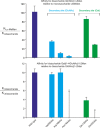

Similar articles
-
Human C-type lectin domain family 4, member C (CLEC4C/BDCA-2/CD303) is a receptor for asialo-galactosyl-oligosaccharides.J Biol Chem. 2011 Oct 14;286(41):35329-35333. doi: 10.1074/jbc.C111.290494. Epub 2011 Aug 31. J Biol Chem. 2011. PMID: 21880719 Free PMC article.
-
Identification of serum glycoprotein ligands for the immunomodulatory receptor blood dendritic cell antigen 2.Glycobiology. 2018 Aug 1;28(8):592-600. doi: 10.1093/glycob/cwy050. Glycobiology. 2018. PMID: 29796630 Free PMC article.
-
Structural analysis of carbohydrate binding by the macrophage mannose receptor CD206.J Biol Chem. 2021 Jan-Jun;296:100368. doi: 10.1016/j.jbc.2021.100368. Epub 2021 Feb 2. J Biol Chem. 2021. PMID: 33545173 Free PMC article.
-
Sialoadhesin and related cellular recognition molecules of the immunoglobulin superfamily.Biochem Soc Trans. 1996 Feb;24(1):150-6. doi: 10.1042/bst0240150. Biochem Soc Trans. 1996. PMID: 8674645 Review. No abstract available.
-
Signaling and immune regulatory role of the dendritic cell immunoreceptor (DCIR) family lectins: DCIR, DCAR, dectin-2 and BDCA-2.Immunobiology. 2004;209(1-2):179-90. doi: 10.1016/j.imbio.2004.03.004. Immunobiology. 2004. PMID: 15481152 Review.
Cited by
-
A robust mRNA signature obtained via recursive ensemble feature selection predicts the responsiveness of omalizumab in moderate-to-severe asthma.Clin Transl Allergy. 2023 Nov;13(11):e12306. doi: 10.1002/clt2.12306. Clin Transl Allergy. 2023. PMID: 38006387 Free PMC article.
-
The Dectin-1 and Dectin-2 clusters: C-type lectin receptors with fundamental roles in immunity.EMBO Rep. 2024 Dec;25(12):5239-5264. doi: 10.1038/s44319-024-00296-2. Epub 2024 Oct 31. EMBO Rep. 2024. PMID: 39482490 Free PMC article. Review.
-
Plasmacytoid dendritic cells at the forefront of anti-cancer immunity: rewiring strategies for tumor microenvironment remodeling.J Exp Clin Cancer Res. 2024 Jul 17;43(1):196. doi: 10.1186/s13046-024-03121-9. J Exp Clin Cancer Res. 2024. PMID: 39020402 Free PMC article. Review.
-
Inhibitory receptors of plasmacytoid dendritic cells as possible targets for checkpoint blockade in cancer.Front Immunol. 2024 Mar 5;15:1360291. doi: 10.3389/fimmu.2024.1360291. eCollection 2024. Front Immunol. 2024. PMID: 38504978 Free PMC article. Review.
-
Mechanism of pathogen recognition by human dectin-2.J Biol Chem. 2017 Aug 11;292(32):13402-13414. doi: 10.1074/jbc.M117.799080. Epub 2017 Jun 26. J Biol Chem. 2017. PMID: 28652405 Free PMC article.
References
-
- Dzionek A., Fuchs A., Schmidt P., Cremer S., Zysk M., Miltenyi S., Buck D. W., Schmitz J. (2000) BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. 165, 6037–6046 - PubMed
-
- Schraml B. U., Reis e Sousa C. (2015) Defining dendritic cells. Curr. Opin. Immunol. 32, 13–20 - PubMed
-
- Weis W. I., Taylor M. E., Drickamer K. (1998) The C-type lectin superfamily in the immune system. Immunol. Rev. 163, 19–34 - PubMed
-
- Guo Y., Feinberg H., Conroy E., Mitchell D. A., Alvarez R., Blixt O., Taylor M. E., Weis W. I., Drickamer K. (2004) Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 11, 591–598 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

