Microarray expression profiling of dysregulated long non-coding RNAs in triple-negative breast cancer
- PMID: 25996380
- PMCID: PMC4622574
- DOI: 10.1080/15384047.2015.1040957
Microarray expression profiling of dysregulated long non-coding RNAs in triple-negative breast cancer
Abstract
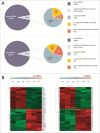
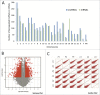
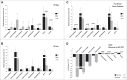
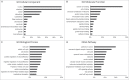
Triple-negative breast cancer (TNBC) represents a collection of malignant breast tumors that are often aggressive and have an increased risk of metastasis and relapse. Long non-coding RNAs are generally defined as RNA transcripts measuring 200 nucleotides or longer that do not encode for any protein. During the past decade, increasing evidence has shown that lncRNAs play important roles in oncogenesis and tumor suppression; however, the roles of lncRNAs in TNBC are poorly understood. To address this issue, we used Agilent human lncRNA microarray chips and bioinformatics tools, including Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG), to assess lncRNA expression in 3 pairs of TNBC tissues. A dysregulated lncRNA expression profile was identified by microarray and verified by qRT-PCR in 48 pairs of breast cancer subtype tissues. Metastasis is the major cause of cancer-related deaths, including those in TNBC, and the presence of dormant residual disseminated tumor cells (DTC) may be a key factor leading to metastasis. ANKRD30A, a potential target for breast cancer immunotherapy, is currently one of the most used DTC markers. Notably, we found the expression levels of the novel intergenic lncRNA LINC00993 to be associated with the expression levels of ANKRD30A. Furthermore, our qRT-PCR data indicated that the expression of LINC00993 was also associated with the expression of the estrogen receptor. In conclusion, our study identified a set of lncRNAs that were consistently aberrantly expressed in TNBC, and these dysregulated lncRNAs may be involved in the development and/or progression of TNBC.
Keywords: ANKRD30A; LINC00993; expression profile; long non-coding RNA; microarray; triple-negative breast cancer.
Figures
References
-
- Polyak K. Heterogeneity in breast cancer. J Clin Invest 2011; 121:3786-8; PMID:21965334; http://dx.doi.org/ 10.1172/JCI60534 - DOI - PMC - PubMed
-
- Bosch A, Eroles P, Zaragoza R, Vina JR, Lluch A. Triple-negative breast cancer: molecular features, pathogenesis, treatment and current lines of research. Cancer Treat Rev 2010; 36:206-15; PMID:20060649; http://dx.doi.org/ 10.1016/j.ctrv.2009.12.002 - DOI - PubMed
-
- Jiao Q, Wu A, Shao G, Peng H, Wang M, Ji S, Liu P, Zhang J. The latest progress in research on triple negative breast cancer (TNBC): risk factors, possible therapeutic targets and prognostic markers. J Thorac Dis 2014; 6:1329-35; PMID:25276378; http://dx.doi.org/ 10.3978/j.issn.2072-1439.2014.08.13 - DOI - PMC - PubMed
-
- Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: review. Pathology 2009; 41:40-7; PMID:19089739; http://dx.doi.org/ 10.1080/00313020802563510 - DOI - PubMed
-
- Lara-Medina F, Perez-Sanchez V, Saavedra-Perez D, Blake-Cerda M, Arce C, Motola-Kuba D, Villarreal-Garza C, Gonzalez-Angulo AM, Bargallo E, Aguilar JL, et al.. Triple-negative breast cancer in Hispanic patients: high prevalence, poor prognosis, and association with menopausal status, body mass index, and parity. Cancer 2011; 117:3658-69; PMID:21387260; http://dx.doi.org/ 10.1002/cncr.25961 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
