Myopathic lamin mutations cause reductive stress and activate the nrf2/keap-1 pathway
- PMID: 25996830
- PMCID: PMC4440730
- DOI: 10.1371/journal.pgen.1005231
Myopathic lamin mutations cause reductive stress and activate the nrf2/keap-1 pathway
Abstract
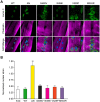
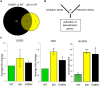
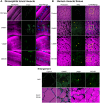
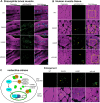
Mutations in the human LMNA gene cause muscular dystrophy by mechanisms that are incompletely understood. The LMNA gene encodes A-type lamins, intermediate filaments that form a network underlying the inner nuclear membrane, providing structural support for the nucleus and organizing the genome. To better understand the pathogenesis caused by mutant lamins, we performed a structural and functional analysis on LMNA missense mutations identified in muscular dystrophy patients. These mutations perturb the tertiary structure of the conserved A-type lamin Ig-fold domain. To identify the effects of these structural perturbations on lamin function, we modeled these mutations in Drosophila Lamin C and expressed the mutant lamins in muscle. We found that the structural perturbations had minimal dominant effects on nuclear stiffness, suggesting that the muscle pathology was not accompanied by major structural disruption of the peripheral nuclear lamina. However, subtle alterations in the lamina network and subnuclear reorganization of lamins remain possible. Affected muscles had cytoplasmic aggregation of lamins and additional nuclear envelope proteins. Transcription profiling revealed upregulation of many Nrf2 target genes. Nrf2 is normally sequestered in the cytoplasm by Keap-1. Under oxidative stress Nrf2 dissociates from Keap-1, translocates into the nucleus, and activates gene expression. Unexpectedly, biochemical analyses revealed high levels of reducing agents, indicative of reductive stress. The accumulation of cytoplasmic lamin aggregates correlated with elevated levels of the autophagy adaptor p62/SQSTM1, which also binds Keap-1, abrogating Nrf2 cytoplasmic sequestration, allowing Nrf2 nuclear translocation and target gene activation. Elevated p62/SQSTM1 and nuclear enrichment of Nrf2 were identified in muscle biopsies from the corresponding muscular dystrophy patients, validating the disease relevance of our Drosophila model. Thus, novel connections were made between mutant lamins and the Nrf2 signaling pathway, suggesting new avenues of therapeutic intervention that include regulation of protein folding and metabolism, as well as maintenance of redox homoeostasis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Suppression of myopathic lamin mutations by muscle-specific activation of AMPK and modulation of downstream signaling.Hum Mol Genet. 2019 Feb 1;28(3):351-371. doi: 10.1093/hmg/ddy332. Hum Mol Genet. 2019. PMID: 30239736 Free PMC article.
-
Nuclear lamins and progerin are dispensable for antioxidant Nrf2 response to arsenic and cadmium.Cell Signal. 2017 May;33:69-78. doi: 10.1016/j.cellsig.2017.02.012. Epub 2017 Feb 14. Cell Signal. 2017. PMID: 28229933 Free PMC article.
-
LMNA variants cause cytoplasmic distribution of nuclear pore proteins in Drosophila and human muscle.Hum Mol Genet. 2012 Apr 1;21(7):1544-56. doi: 10.1093/hmg/ddr592. Epub 2011 Dec 20. Hum Mol Genet. 2012. PMID: 22186027 Free PMC article.
-
Laminopathies: what can humans learn from fruit flies.Cell Mol Biol Lett. 2018 Jul 6;23:32. doi: 10.1186/s11658-018-0093-1. eCollection 2018. Cell Mol Biol Lett. 2018. PMID: 30002683 Free PMC article. Review.
-
Lamins as mediators of oxidative stress.Biochem Biophys Res Commun. 2012 May 18;421(4):635-9. doi: 10.1016/j.bbrc.2012.04.058. Epub 2012 Apr 17. Biochem Biophys Res Commun. 2012. PMID: 22538370 Review.
Cited by
-
Phosphorylated TDP-43 (pTDP-43) aggregates in the axial skeletal muscle of patients with sporadic and familial amyotrophic lateral sclerosis.Acta Neuropathol Commun. 2018 Apr 13;6(1):28. doi: 10.1186/s40478-018-0528-y. Acta Neuropathol Commun. 2018. PMID: 29653597 Free PMC article.
-
Responses to reductive stress in the cardiovascular system.Free Radic Biol Med. 2017 Aug;109:114-124. doi: 10.1016/j.freeradbiomed.2016.12.006. Epub 2016 Dec 8. Free Radic Biol Med. 2017. PMID: 27940350 Free PMC article. Review.
-
Forced expression of miR-143 and -145 in cardiomyocytes induces cardiomyopathy with a reductive redox shift.Cell Mol Biol Lett. 2020 Aug 24;25:40. doi: 10.1186/s11658-020-00232-x. eCollection 2020. Cell Mol Biol Lett. 2020. PMID: 32855642 Free PMC article.
-
Cellular and Animal Models of Striated Muscle Laminopathies.Cells. 2019 Mar 29;8(4):291. doi: 10.3390/cells8040291. Cells. 2019. PMID: 30934932 Free PMC article. Review.
-
Drosophila p38 MAPK interacts with BAG-3/starvin to regulate age-dependent protein homeostasis.Aging Cell. 2021 Nov;20(11):e13481. doi: 10.1111/acel.13481. Epub 2021 Oct 21. Aging Cell. 2021. PMID: 34674371 Free PMC article.
References
-
- Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U (2007) Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol 8: 562–573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

