Evaluation of Brachypodium distachyon L-Tyrosine Decarboxylase Using L-Tyrosine Over-Producing Saccharomyces cerevisiae
- PMID: 25996877
- PMCID: PMC4440718
- DOI: 10.1371/journal.pone.0125488
Evaluation of Brachypodium distachyon L-Tyrosine Decarboxylase Using L-Tyrosine Over-Producing Saccharomyces cerevisiae
Abstract
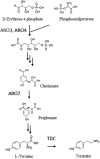
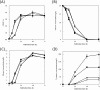
To demonstrate that herbaceous biomass is a versatile gene resource, we focused on the model plant Brachypodium distachyon, and screened the B. distachyon for homologs of tyrosine decarboxylase (TDC), which is involved in the modification of aromatic compounds. A total of 5 candidate genes were identified in cDNA libraries of B. distachyon and were introduced into Saccharomyces cerevisiae to evaluate TDC expression and tyramine production. It is suggested that two TDCs encoded in the transcripts Bradi2g51120.1 and Bradi2g51170.1 have L-tyrosine decarboxylation activity. Bradi2g51170.1 was introduced into the L-tyrosine over-producing strain of S. cerevisiae that was constructed by the introduction of mutant genes that promote deregulated feedback inhibition. The amount of tyramine produced by the resulting transformant was 6.6-fold higher (approximately 200 mg/L) than the control strain, indicating that B. distachyon TDC effectively converts L-tyrosine to tyramine. Our results suggest that B. distachyon possesses enzymes that are capable of modifying aromatic residues, and that S. cerevisiae is a suitable host for the production of L-tyrosine derivatives.
Conflict of interest statement
Figures

Similar articles
-
Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice.Planta. 2007 Dec;227(1):263-72. doi: 10.1007/s00425-007-0614-z. Epub 2007 Sep 1. Planta. 2007. PMID: 17763868
-
Characterization of the tyramine-producing pathway in Sporolactobacillus sp. P3J.Microbiology (Reading). 2011 Jun;157(Pt 6):1841-1849. doi: 10.1099/mic.0.046367-0. Epub 2011 Mar 17. Microbiology (Reading). 2011. PMID: 21415114
-
First evidence of a membrane-bound, tyramine and beta-phenylethylamine producing, tyrosine decarboxylase in Enterococcus faecalis: a two-dimensional electrophoresis proteomic study.Proteomics. 2009 May;9(10):2695-710. doi: 10.1002/pmic.200800780. Proteomics. 2009. PMID: 19405032
-
Tyramine and phenylethylamine biosynthesis by food bacteria.Crit Rev Food Sci Nutr. 2012;52(5):448-67. doi: 10.1080/10408398.2010.500545. Crit Rev Food Sci Nutr. 2012. PMID: 22369263 Review.
-
Brachypodium distachyon as a Genetic Model System.Annu Rev Genet. 2015;49:1-20. doi: 10.1146/annurev-genet-112414-055135. Epub 2015 Sep 17. Annu Rev Genet. 2015. PMID: 26393966 Review.
Cited by
-
Functional characterization of tyrosine decarboxylase genes that contribute to acteoside biosynthesis in Rehmannia glutinosa.Planta. 2022 Feb 11;255(3):64. doi: 10.1007/s00425-022-03849-8. Planta. 2022. PMID: 35147783
-
Biosensor and machine learning-aided engineering of an amaryllidaceae enzyme.Nat Commun. 2024 Mar 7;15(1):2084. doi: 10.1038/s41467-024-46356-y. Nat Commun. 2024. PMID: 38453941 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

